

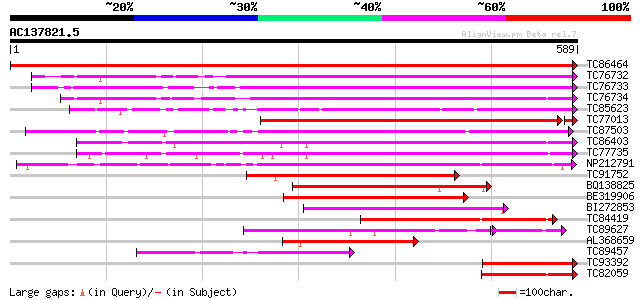
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.5 - phase: 0
(589 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 1171 0.0
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 444 e-125
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 428 e-120
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 419 e-117
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 417 e-117
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 407 e-114
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 397 e-111
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 375 e-104
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 362 e-100
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 328 4e-90
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 252 2e-67
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 223 1e-58
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 209 3e-54
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 198 6e-51
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 183 2e-46
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 147 1e-45
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 159 4e-39
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 144 7e-35
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 129 2e-30
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 128 7e-30
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 1171 bits (3029), Expect = 0.0
Identities = 588/589 (99%), Positives = 588/589 (99%)
Frame = +1
Query: 1 MNKIKQSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSL 60
MNKIKQSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSL
Sbjct: 49 MNKIKQSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSL 228
Query: 61 SHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTV 120
SHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTV
Sbjct: 229 SHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTV 408
Query: 121 EKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISS 180
EKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISS
Sbjct: 409 EKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISS 588
Query: 181 AITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV 240
AITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV
Sbjct: 589 AITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV 768
Query: 241 LGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAP 300
LGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAP
Sbjct: 769 LGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAP 948
Query: 301 LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVA 360
LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVA
Sbjct: 949 LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVA 1128
Query: 361 IVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNC 420
IVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNC
Sbjct: 1129IVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNC 1308
Query: 421 FISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 480
FISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK
Sbjct: 1309FISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 1488
Query: 481 DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTG 540
DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTG
Sbjct: 1489DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTG 1668
Query: 541 PGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
PGAGTSKRVTWKGFKVITSAAEAQS TPGNFIGGSSWLGSTGFPFSLGL
Sbjct: 1669PGAGTSKRVTWKGFKVITSAAEAQSSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 444 bits (1141), Expect = e-125
Identities = 262/583 (44%), Positives = 351/583 (59%), Gaps = 16/583 (2%)
Frame = +2
Query: 23 SLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYPELCF 82
S+ T +V S+VAI I+SS+L +SH + I S + P LC
Sbjct: 74 SIPKTFWLVLSLVAI---------------ISSSALIISHLNKPI--SFFNLSSAPNLCE 202
Query: 83 SAISSEPNITH--------KITNHKD-VISLSLNITTRAVEH--NYFTVEKLLLRKSLTK 131
A+ ++ +TH ++N KD +S +++ T++ H ++ R+ +
Sbjct: 203 HALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSP 382
Query: 132 REKIALHDCLETIDETLDELKEAQNDLVLYPSKKTL-YQHADDLKTLISSAITNQVTCLD 190
RE+IAL+DC E +D ++D + D VL +K + QH D T +SS +TN TCLD
Sbjct: 383 REEIALNDCEELMDLSMDRVW----DSVLTLTKNNIDSQH--DAHTWLSSVLTNHATCLD 544
Query: 191 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLL 250
G +V+ E +H ++ + + VL N +
Sbjct: 545 GLEGSS------RVVMESDLH-------------DLISRARSSLAVLVSVLPPKANDGFI 667
Query: 251 EEENGVGWPEWISAGDRRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYV 308
+E+ +P W+++ DRRLL+ S +KA+VVVA DGSG FKTV++AVA+AP RYV
Sbjct: 668 DEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYV 847
Query: 309 IKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLA 368
I +K G YKEN+E+ KKKTN+M +GDG TIITGS N +DG+TTF SATVA VG F+A
Sbjct: 848 IYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIA 1027
Query: 369 RDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDF 428
+DI FQNTAGP KHQAVALRVGAD S C I A+QDTLY H+NRQF+ + +I+GTVDF
Sbjct: 1028QDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDF 1207
Query: 429 IFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGN 488
IFGN+AVVFQ + AR+P + QKNMVTAQGR DPNQNT IQ+C + + DL+ V+G+
Sbjct: 1208IFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGS 1387
Query: 489 FPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWN--GNFALNTLVYREYQNTGPGAGTS 546
TYLGRPWK+YSRTV +QS + IDP GW EW+ L TL Y EY N+G GAGT
Sbjct: 1388IKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTG 1567
Query: 547 KRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
KRVTW G+ +I +AAEA FT I G+ WL +TG F GL
Sbjct: 1568KRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 1696
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 428 bits (1101), Expect = e-120
Identities = 248/574 (43%), Positives = 339/574 (58%), Gaps = 7/574 (1%)
Frame = +2
Query: 23 SLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYPELCF 82
SL T ++ S+VAI+ ++ S K +S H + + C + + C
Sbjct: 86 SLPKTFWLILSLVAIIISSALISTHLKK------PISFFHLT-TVQNVYCEHAVDTKSCL 244
Query: 83 SAISSEPNITHKITNHKDVISLSLNITTRAVEH--NYFTVEKLLLRKSLTKREKIALHDC 140
+ +S ++ +T + + L++ T++ H N ++ R+ + RE+IAL DC
Sbjct: 245 AHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRRINSPREEIALSDC 424
Query: 141 LETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKE 200
+ +D +++ + + L K D T +SS +TN TCLDG
Sbjct: 425 EQLMDLSMNRIWDTMLKLT-----KNNIDSQQDAHTWLSSVLTNHATCLDGL-------- 565
Query: 201 VRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVG-WP 259
EG V + N L + +A F +V+ K+R +E +G +P
Sbjct: 566 ------EGSSRV--VMENDLQDLISRARSSLAVF----LVVFPQKDRDQFIDETLIGEFP 709
Query: 260 EWISAGDRRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYK 317
W+++ DRRLL+ + +KA+VVVA DGSG FKTV+EAVA+AP +YVI +K G YK
Sbjct: 710 SWVTSKDRRLLETAVGDIKANVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKGTYK 889
Query: 318 ENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTA 377
ENVE+ KKTN+M +GDG TIITG+ N +DG+TTF S+TVA VG F+A+DI FQN A
Sbjct: 890 ENVEIGSKKTNVMLVGDGMDATIITGNLNFIDGTTTFKSSTVAAVGDGFIAQDIWFQNMA 1069
Query: 378 GPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVF 437
G AKHQAVALRVG+D S C I A+QDTLY H+NRQF+ + I+GT+DFIFGN+AVVF
Sbjct: 1070GAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAAVVF 1249
Query: 438 QNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPW 497
Q C + AR+P + Q NM TAQGR DP QNTG IQ+C + + DL+ V G+ T+LGRPW
Sbjct: 1250QKCKLVARKPMANQNNMFTAQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLGRPW 1429
Query: 498 KEYSRTVFMQSSISDVIDPVGWHEWN--GNFALNTLVYREYQNTGPGAGTSKRVTWKGFK 555
K+YSRTV MQS + IDP GW EW+ L TL Y EY N GPGAGT+KRVTW G+
Sbjct: 1430KKYSRTVVMQSFLDSHIDPTGWAEWDAASKDFLQTLYYGEYLNNGPGAGTAKRVTWPGYH 1609
Query: 556 VITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
VI +AAEA FT I G+ WL +TG F+ GL
Sbjct: 1610VINTAAEASKFTVAQLIQGNVWLKNTGVAFTEGL 1711
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 419 bits (1078), Expect = e-117
Identities = 248/554 (44%), Positives = 333/554 (59%), Gaps = 17/554 (3%)
Frame = +3
Query: 53 IASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITH--------KITNHKD-VIS 103
I+SS+L +SH + I S + P +C A+ + +TH + N KD +S
Sbjct: 126 ISSSALIISHLNKPI--SIFHFSSAPNVCEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLS 299
Query: 104 LSLNITTRAVEH--NYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLY 161
+++ T++ H ++ R+ + RE+ AL+ C + ++ +++ + D VL
Sbjct: 300 TLISLLTKSTTHIRKAMDTANVIKRRINSPREENALNVCEKLMNLSMERVW----DSVLT 467
Query: 162 PSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIH--VEHMCSNA 219
+K + D T +SS +TN TCLDG + + + E I + S+
Sbjct: 468 LTKDNM-DSQQDAHTWLSSVLTNHATCLDGL------EGTSRAVMENDIQDLIARARSSL 626
Query: 220 LAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGST--VKA 277
+ + KD EF ++E +P W+++ DRRLL+ S VKA
Sbjct: 627 AVLVAVLPPKDHDEF---------------IDESLNGDFPSWVTSKDRRLLESSVGDVKA 761
Query: 278 DVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRT 337
+VVVA DGSG FKTV+EAVA+AP K + RYVI +K G+YKENVE+ KTN+M LGDG
Sbjct: 762 NVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKKGIYKENVEIASSKTNVMLLGDGMD 941
Query: 338 NTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFY 397
TIITGS N VDG+ TF +ATVA VG F+A+DI FQNTAGP KHQAVALRVG+D S
Sbjct: 942 ATIITGSLNYVDGTGTFQTATVAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVIN 1121
Query: 398 NCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTA 457
C I A+QDTLY H NRQF+ + FI+GT+DFIFG++AVV Q C + AR+P + Q NMVTA
Sbjct: 1122RCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGDAAVVLQKCKLVARKPMANQNNMVTA 1301
Query: 458 QGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPV 517
QGR+DPNQNT IQ+C + + DL+ V G+ TYLGRPWK+YSRTV MQS + IDP
Sbjct: 1302QGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDPT 1481
Query: 518 GWHEWN--GNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGS 575
GW EW+ L TL Y EY N+GPGAGTSKRV W G+ +I + AEA FT I G+
Sbjct: 1482GWAEWDAASKDFLQTLYYGEYMNSGPGAGTSKRVKWPGYHII-NTAEANKFTVAQLIQGN 1658
Query: 576 SWLGSTGFPFSLGL 589
WL +TG F GL
Sbjct: 1659VWLKNTGVAFIAGL 1700
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 417 bits (1072), Expect = e-117
Identities = 247/532 (46%), Positives = 321/532 (59%), Gaps = 5/532 (0%)
Frame = +3
Query: 63 HSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAV---EHNYFT 119
+S + +KS C+ T YP+ C +++ I + D +SL I E N
Sbjct: 636 YSLSSIKSWCSQTPYPQPCEYYLTNNA-FNQTIKSKSDFFKVSLEIAMERAKKGEENTHA 812
Query: 120 VEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLIS 179
V K + +EK A DCLE D T+ +L + + +K T Y+ +T +S
Sbjct: 813 VGP----KCRSPQEKAAWADCLELYDFTVQKLSQTKY------TKCTQYES----QTWLS 950
Query: 180 SAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNM 239
+A+TN TC +GF V +L +V + SN L++ K ++Q
Sbjct: 951 TALTNLETCKNGFYDLGVTNYVLPLLSN---NVTKLLSNTLSLNK-------VPYQQP-- 1094
Query: 240 VLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTV--KADVVVAADGSGNFKTVSEAVA 297
S K+ G+P W+ GDR+LLQ S+ KA+VVVA DGSG + TV A
Sbjct: 1095 ---SYKD----------GFPTWVKPGDRKLLQTSSAASKANVVVAKDGSGKYTTVKAATD 1235
Query: 298 AAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSA 357
AAP S RYVI +KAGVY E VE+ K N+M +GDG TIITGS++V G+TTF SA
Sbjct: 1236 AAP-SGSGRYVIYVKAGVYNEQVEI--KAKNVMLVGDGIGKTIITGSKSVGGGTTTFRSA 1406
Query: 358 TVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFF 417
TVA G F+A+DITF+NTAG A HQAVA R G+DLS FY C YQDTLYVH+ RQF+
Sbjct: 1407 TVAATGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFY 1586
Query: 418 VNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIG 477
C I GTVDFIFGN+AVV QNC+I AR P + VTAQGR DPNQNTGI+I R+
Sbjct: 1587 RECNIYGTVDFIFGNAAVVLQNCNIFARNP-PAKTITVTAQGRTDPNQNTGIIIHNSRVS 1763
Query: 478 ATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQ 537
A DL + +YLGRPW++YSRTVFM++ + I+P GW W+GNFAL+TL Y EY
Sbjct: 1764 AQSDLN--PSSVKSYLGRPWQKYSRTVFMKTVLDGFINPAGWLPWDGNFALDTLYYAEYA 1937
Query: 538 NTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
NTG G+ TS RVTWKG+ V+TSA++A FT GNFI G+SW+G+TG PF+ GL
Sbjct: 1938 NTGSGSSTSNRVTWKGYHVLTSASQASPFTVGNFIAGNSWIGNTGVPFTSGL 2093
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 407 bits (1046), Expect(2) = e-114
Identities = 197/316 (62%), Positives = 242/316 (76%), Gaps = 2/316 (0%)
Frame = +2
Query: 261 WISAGDRRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKE 318
W+S DR+LLQ + K D+VVA DG+GNF T+ EA+AAAP S+ R+VI IKAG Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 319 NVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAG 378
NVEV KKKTN+M +GDG T++ SRNVVDG TTF S+T A+VG F+A+ ITF+N+AG
Sbjct: 221 NVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFENSAG 400
Query: 379 PAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQ 438
P+KHQAVA+R GAD SAFY C +AYQDTLYVH+ RQF+ C + GTVDFIFGN+AVVFQ
Sbjct: 401 PSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVFQ 580
Query: 439 NCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWK 498
NC+++AR+P+ QKN+ TAQGR DPNQNTGI I C+I A DL V+ F +YLGRPWK
Sbjct: 581 NCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRPWK 760
Query: 499 EYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVIT 558
+YSRTV++ S I ++IDP GW EWNG FAL+TL Y EY+N GPG+ TS RVTW G+KVIT
Sbjct: 761 KYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYKVIT 940
Query: 559 SAAEAQSFTPGNFIGG 574
+A EA FT FI G
Sbjct: 941 NATEASQFTVRQFIQG 988
Score = 22.7 bits (47), Expect(2) = e-114
Identities = 9/13 (69%), Positives = 9/13 (69%)
Frame = +1
Query: 577 WLGSTGFPFSLGL 589
WL STG PF L L
Sbjct: 997 WLNSTGVPFFLDL 1035
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 397 bits (1020), Expect = e-111
Identities = 230/579 (39%), Positives = 336/579 (57%), Gaps = 10/579 (1%)
Frame = +2
Query: 17 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 76
+K++ I + +++++VA I+A VA + + ++++S +S + LK+ C +T
Sbjct: 134 RKRITIIIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQ 313
Query: 77 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIA 136
YP CFS+ISS P+ T+ + + LSL + + T K+ R K A
Sbjct: 314 YPNSCFSSISSLPD--SNTTDPEQLFKLSLKVAIDELSKLSLT---RFSEKATEPRVKKA 478
Query: 137 LHDCLETIDETLDELKEAQNDLV-----LYPSKKTLYQHADDLKTLISSAITNQVTCLD- 190
+ C + ++LD L ++ + +V L P+K D++T +S+A+T+ TCLD
Sbjct: 479 IGVCDNVLADSLDRLNDSMSTIVDGGKMLSPAK------IRDVETWLSAALTDHDTCLDA 640
Query: 191 -GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKL 249
G + A + V ++ + SN+LA+ + + ++ FE SN +R+L
Sbjct: 641 VGEVNSTAARGVIPEIERIMRNSTEFASNSLAIVSKVI-RLLSNFEV------SNHHRRL 799
Query: 250 LEEENGVGWPEWISAGDRRLLQG--STVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRY 307
L E +PEW+ +RRLL + D VVA DGSG +KT+ EA+ KS +R+
Sbjct: 800 LGE-----FPEWLGTAERRLLATVVNETVPDAVVAKDGSGQYKTIGEALKLVKKKSLQRF 964
Query: 308 VIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFL 367
V+ +K GVY EN+++ K N+M GDG T T+++GSRN +DG+ TF +AT A+ G F+
Sbjct: 965 VVYVKKGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGKGFI 1144
Query: 368 ARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVD 427
A+DI F NTAG +KHQAVA+R G+D S FY C + YQDTLY H+NRQF+ +C I+GT+D
Sbjct: 1145AKDIQFLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTID 1324
Query: 428 FIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKG 487
FIFGN+A VFQNC I R+P S Q N +TAQG+ DPNQN+GIVIQK L G
Sbjct: 1325FIFGNAAAVFQNCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQK---STFTTLPGDNL 1495
Query: 488 NFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNF-ALNTLVYREYQNTGPGAGTS 546
PTYLGRPWK++S T+ M+S I + PVGW W N ++++Y EYQNTGPGA
Sbjct: 1496IAPTYLGRPWKDFSTTIIMKSEIGSFLKPVGWISWVANVEPPSSILYAEYQNTGPGADVP 1675
Query: 547 KRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPF 585
RV W G+K +A FT +FI G WL S F
Sbjct: 1676GRVKWAGYKPALGDEDAIKFTVDSFIQGPEWLPSASVQF 1792
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 375 bits (962), Expect = e-104
Identities = 226/535 (42%), Positives = 306/535 (56%), Gaps = 15/535 (2%)
Frame = +3
Query: 70 SACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSL 129
+AC +T P C S + + + SLS + + + Y L R +L
Sbjct: 132 AACKSTPDPTYCKSILPPQNANVYDYGRFSVKKSLSQSRKFSDLINKY-----LQRRSTL 296
Query: 130 TKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQ---HADDLKTLISSAITNQV 186
+ AL DC D D L + + +K T + ++++TL+S+ +TNQ
Sbjct: 297 STTALRALQDCQSLSDLNFDFLSSSFQTV----NKTTKFLPSLQGENIQTLLSAILTNQQ 464
Query: 187 TCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAM-TKNMTDKDIAEFEQTNMVLGSN- 244
TCLDG + R L + + S +LA TK + + N +
Sbjct: 465 TCLDGLKDTSSAWSFRNGLTIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSFPNSKHSNKG 644
Query: 245 -KNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVV------AADGSGNFKTVSEAVA 297
KN +L + + S R+LLQ + V DVVV + DGSGNF T+++A+A
Sbjct: 645 FKNGRLPLKMTSKTRAIYESVSRRKLLQSNQVGEDVVVRDIVTVSQDGSGNFTTINDAIA 824
Query: 298 AAPLKSSKR---YVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTF 354
AAP KS ++I + AGVY+E + + KKKT +M +GDG TIITG+ +VVDG TTF
Sbjct: 825 AAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDGINKTIITGNHSVVDGWTTF 1004
Query: 355 HSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNR 414
S T A+VG F+ ++T +NTAG KHQAVALR GADLS FY+C YQDTLY H+ R
Sbjct: 1005GSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQDTLYTHSLR 1184
Query: 415 QFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKC 474
QF+ C I GTVDFIFGN+ VVFQNC+++ R P SGQ N +TAQGR DPNQ+TG I C
Sbjct: 1185QFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAITAQGRTDPNQDTGTSIHNC 1364
Query: 475 RIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYR 534
I AT DL G TYLGRPWKEYSRTV+MQ+ + +VI+ GW W+G FAL+TL Y
Sbjct: 1365TIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGEFALSTLYYA 1544
Query: 535 EYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
E+ N+GPG+ T RVTW+G+ VI +A +A +FT NF+ G WL TG ++ L
Sbjct: 1545EFNNSGPGSSTDGRVTWQGYHVI-NATDAANFTVANFLLGDDWLPQTGVSYTNSL 1706
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 362 bits (928), Expect = e-100
Identities = 225/550 (40%), Positives = 310/550 (55%), Gaps = 30/550 (5%)
Frame = +2
Query: 70 SACTTTLYPELC---FSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLR 126
+AC TTLYP+LC SAI S P+ + K I +L + + + + +
Sbjct: 149 AACKTTLYPKLCRSMLSAIRSSPSDPYNYG--KFSIKQNLKVARKLEKVFIDFLNRHQSS 322
Query: 127 KSLTKREKIALHDC-------LETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLIS 179
SL E AL DC ++ ++ DELK A + S + + D +++ +S
Sbjct: 323 SSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSS-----SSSSDTELVDKIESYLS 487
Query: 180 SAITNQVTCLDGF---SHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQ 236
+ TN TC DG + A+ + Q + + A++KNM +
Sbjct: 488 AVATNHYTCYDGLVVTKSNIANALAVPLKDATQFYSVSLGLVTEALSKNMKRNKTRKHGL 667
Query: 237 TNMVLGSNKNRKLLEEENGVGWPEW---------ISAGDRRLLQ-----GSTVKADVVVA 282
N S K R+ LE+ + ++ S R+L+ G + V+V+
Sbjct: 668 PNK---SFKVRQPLEKLIKLLRTKYSCQKTSSNCTSTRTERILKESESHGILLNDFVLVS 838
Query: 283 ADGSGNFKTVSEAVAAAPLKSSKR---YVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNT 339
G N ++ +A+AAAP + Y+I ++ G Y+E V VPK K NI+ +GDG NT
Sbjct: 839 PYGIANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGINNT 1018
Query: 340 IITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNC 399
IITG+ +V+DG TTF+S+T A+ G F+A DITF+NTAGP KHQAVA+R ADLS FY C
Sbjct: 1019IITGNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRC 1198
Query: 400 DIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQG 459
YQDTLYVH+ RQF+ +C I GTVDFIFGN+AVVFQNC+I+AR+P QKN VTAQG
Sbjct: 1199SFEGYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAVTAQG 1378
Query: 460 RVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGW 519
R DPNQNTGI IQ C I A +DL + +YLGRPWK YSRTV+MQS I D + P GW
Sbjct: 1379RTDPNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGW 1558
Query: 520 HEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLG 579
EWNG L+T+ Y E+ N GPG+ T+ RV W G ++ +A +FT NF G++WL
Sbjct: 1559LEWNGTVGLDTIFYGEFNNYGPGSVTNNRVQWPGHFLLND-TQAWNFTVLNFTLGNTWLP 1735
Query: 580 STGFPFSLGL 589
T P++ GL
Sbjct: 1736DTDIPYTEGL 1765
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 328 bits (841), Expect = 4e-90
Identities = 209/594 (35%), Positives = 318/594 (53%), Gaps = 13/594 (2%)
Frame = +1
Query: 8 LAGISNSGNK---KKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHS 64
+ G N+G + KK + + +L+VA + + + TK + + I++S ++
Sbjct: 1 MGGNDNNGGQGQGKKHALLGVSCILLVAMVGVVAVSLTKGGDGEQKAHISNSQKNVD--- 171
Query: 65 HAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEH--NYFTVEK 122
C +T + E C + +N K+ I +L T + N + +
Sbjct: 172 -----MLCQSTKFKETCHKTLEKA-----SFSNMKNRIKGALGATEEELRKHINNSALYQ 321
Query: 123 LLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAI 182
L S+TK+ A+ C E +D +D + ++ L + K L ++A D+K ++ +
Sbjct: 322 ELATDSMTKQ---AMEICNEVLDYAVDGIHKSVGTLDQFDFHK-LSEYAFDIKVWLTGTL 489
Query: 183 TNQVTCLDGF--SHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV 240
++Q TCLDGF + A + + KVL+ + SNA+ M ++ + + F +
Sbjct: 490 SHQQTCLDGFVNTKTHAGETMAKVLKTSM----ELSSNAIDMM-DVVSRILKGFHPSQYG 654
Query: 241 LGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAP 300
+ +R+LL ++ G P W+S G R LL G VKA+ VVA DGSG FKT+++A+ P
Sbjct: 655 V----SRRLLSDD---GIPSWVSDGHRHLLAGGNVKANAVVAQDGSGQFKTLTDALKTVP 813
Query: 301 LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVA 360
++ +VI +KAGVYKE V V K+ + +GDG T T TGS N DG T+ +AT
Sbjct: 814 PTNAAPFVIYVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFG 993
Query: 361 IVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNC 420
+ G NF+A+DI F+NTAG +K QAVALRV AD + F+NC + +QDTL+V + RQF+ +C
Sbjct: 994 VNGANFMAKDIGFENTAGTSKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDC 1173
Query: 421 FISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 480
ISGT+DF+FG++ VFQNC + R P GQK +VTA GR N + +V
Sbjct: 1174AISGTIDFVFGDAFGVFQNCKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEP 1353
Query: 481 DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTG 540
L V +YLGRPWK YS+ V M S+I + P G+ G +T + EY N G
Sbjct: 1354ALTSVTPKL-SYLGRPWKLYSKVVIMDSTIDAMFAPEGYMPMVGGAFKDTCTFYEYNNKG 1530
Query: 541 PGAGTSKRVTWKGFKVITSAAEAQSFTPGNFI------GGSSWLGSTGFPFSLG 588
PGA T+ RV W G KV+TS A+ + PG F +W+ +G P+SLG
Sbjct: 1531PGADTNLRVKWHGVKVLTSNVAAE-YYPGKFFEIVNATARDTWIVKSGVPYSLG 1689
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 252 bits (644), Expect = 2e-67
Identities = 127/224 (56%), Positives = 157/224 (69%), Gaps = 3/224 (1%)
Frame = +3
Query: 247 RKLLEEENGVGWPEWISAGDRRLLQGST---VKADVVVAADGSGNFKTVSEAVAAAPLKS 303
R+LL + P W+ DR+LLQ + KAD+VVA DGSG +KT+S A+ P KS
Sbjct: 18 RRLLSLSHQNEEPNWLHHKDRKLLQKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNKS 197
Query: 304 SKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVG 363
KR VI +K G+Y ENV V K K N+M +GDG TI++GS N +DG+ TF +AT A+ G
Sbjct: 198 DKRTVIYVKKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFG 377
Query: 364 GNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFIS 423
NF+ARDI F+NTAGP KHQAVAL AD + FY C + AYQDTLY H+NRQF+ C I
Sbjct: 378 RNFIARDIGFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIY 557
Query: 424 GTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNT 467
GTVDFIFGNSAVV QNC+I R+P GQ+N +TAQG+ DPN NT
Sbjct: 558 GTVDFIFGNSAVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 223 bits (569), Expect = 1e-58
Identities = 117/218 (53%), Positives = 146/218 (66%), Gaps = 11/218 (5%)
Frame = +1
Query: 294 EAVAAAPLKS--SKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVD-G 350
EAV AAP KR+VI IK GVY+E V VP KK N++FLGDG T+ITGS NV G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 351 STTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYV 410
TT++SATVA++G F+A+D+T +NTAGP HQAVA R+ +DLS NC+ + QDTLY
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 411 HNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHAR----RPNSGQKNMVTAQGRVDPNQN 466
H+ RQF+ +C I G VDFIFGNSA +FQ+C I R +P G+ N +TA GR DP Q+
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 467 TGIVIQKCRIGATKDLEGVKGNFP----TYLGRPWKEY 500
TG V Q C I T+D + + P YLGRPWKEY
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 209 bits (531), Expect = 3e-54
Identities = 107/192 (55%), Positives = 129/192 (66%)
Frame = +2
Query: 285 GSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGS 344
GSG++ V +AV+AAP S KRYVI +K GVY ENVE+ KKK NIM +G+G TII+GS
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 345 RNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAY 404
RN VDGSTTF SAT A+ G F+ARDI+FQNTAG KHQAVALR +DLS FY C I Y
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 405 QDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPN 464
QD+LY H RQF+ C ISGTVDFIFG++ VFQNC I A+ ++ K P
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKERDA*TKEHCYCSRTKRPE 541
Query: 465 QNTGIVIQKCRI 476
++ C I
Sbjct: 542 PTNWFLLPFCNI 577
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 198 bits (503), Expect = 6e-51
Identities = 106/218 (48%), Positives = 132/218 (59%), Gaps = 5/218 (2%)
Frame = +1
Query: 306 RYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGN 365
RY I +KAGVY E + +PK NI+ GDG TI+TG +N G T +AT A
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTALG 183
Query: 366 FLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGT 425
F+ + +TF+NTAGPA HQAVA R D+SA C I+ YQDTLYV NRQF+ NC ISGT
Sbjct: 184 FIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISGT 363
Query: 426 VDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGV 485
VDFIFG SA + Q+ I R P+ Q N +TA G NTGIVIQ C I L
Sbjct: 364 VDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFPQ 543
Query: 486 KGNFPTYLGRPWKEYSRTVFMQSSI-----SDVIDPVG 518
+ +YLGRPWK ++TV M+S+I S +DP+G
Sbjct: 544 RFTIKSYLGRPWKVLTKTVVMESTIG*FYSSXWMDPLG 657
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 183 bits (464), Expect = 2e-46
Identities = 91/205 (44%), Positives = 126/205 (61%)
Frame = +2
Query: 365 NFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISG 424
+F A ++ F+N+AG AKHQAVALRV AD + FYNC + YQDTLY + RQF+ +C I+G
Sbjct: 83 HFTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITG 262
Query: 425 TVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEG 484
T+DF+FG++ VFQNC + R+P + Q+ MVTA GR + + +V Q C ++
Sbjct: 263 TIDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLT 442
Query: 485 VKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAG 544
++ YLGRPW+ +S+ V + S I + P G+ W GN T Y EY N G GA
Sbjct: 443 MQPKI-AYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGAGAA 619
Query: 545 TSKRVTWKGFKVITSAAEAQSFTPG 569
T+ +V W G K I SA EA + PG
Sbjct: 620 TNLKVKWPGVKTI-SAGEAAKYYPG 691
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 147 bits (370), Expect(2) = 1e-45
Identities = 96/273 (35%), Positives = 141/273 (51%), Gaps = 10/273 (3%)
Frame = +3
Query: 244 NKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKS 303
N RK + E+ + D +L + + K + V+ DGS FK+++EA+ + +
Sbjct: 132 NYQRKTIMEKRYRNVSGNVQGLDPKLKKAESNKVRLKVSQDGSAQFKSITEALNSIQPYN 311
Query: 304 SKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGS-RNVVDGST-----TFHSA 357
+R +I I G Y+E + VPK I FLGD R ITG+ V GS TF+SA
Sbjct: 312 IRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPTITGNDTQSVTGSDGAQLRTFNSA 491
Query: 358 TVAIVGGNFLARDITFQNTA----GPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNN 413
TVA+ F+A +I F+NTA G QAVA+R+ + +AFYNC QDTLY H
Sbjct: 492 TVAVNASYFMAININFENTASFPIGSKVEQAVAVRITGNKTAFYNCTFSGVQDTLYDHKG 671
Query: 414 RQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQK 473
+F NC I G+VDFI G+ +++ C I R + +TAQ +P+ ++G +
Sbjct: 672 LHYFNNCTIKGSVDFICGHGKSLYEGCTI---RSIANNMTSITAQSGSNPSYDSGFSFKN 842
Query: 474 CRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFM 506
V G+ PTYLGRPW + + F+
Sbjct: 843 SM---------VIGDGPTYLGRPWGKLFTSCFL 914
Score = 55.1 bits (131), Expect(2) = 1e-45
Identities = 27/79 (34%), Positives = 42/79 (52%)
Frame = +1
Query: 500 YSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITS 559
YS+ VF + + + + P GW +WN Y EY+ +GPG+ T+ RV W + +
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPWAR---MLN 1065
Query: 560 AAEAQSFTPGNFIGGSSWL 578
EAQ F +I G++WL
Sbjct: 1066DKEAQVFIGTQYIDGNTWL 1122
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 159 bits (401), Expect = 4e-39
Identities = 81/144 (56%), Positives = 104/144 (71%), Gaps = 3/144 (2%)
Frame = +1
Query: 284 DGSGNFKTVSEAVAAAP---LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTI 340
DGSGNF T+++A+AAAP + S ++I I GVY+E V + KKK +M +G+G TI
Sbjct: 64 DGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQTI 243
Query: 341 ITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCD 400
ITG+RNV DG TTF+SAT A+V F+A +ITF+NTAG AK+QAVALR GAD+S FY+C
Sbjct: 244 ITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSCS 423
Query: 401 IIAYQDTLYVHNNRQFFVNCFISG 424
YQDTLY H+ RQF+ C I G
Sbjct: 424 FEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 144 bits (364), Expect = 7e-35
Identities = 91/228 (39%), Positives = 127/228 (54%), Gaps = 1/228 (0%)
Frame = +1
Query: 132 REKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHAD-DLKTLISSAITNQVTCLD 190
R A+ DCL+ +D +LD+L ++ + K + DL+T +S+ + TC++
Sbjct: 313 RTSNAVSDCLDLLDMSLDQLNQSISAAQKPKEKDNSTGKLNCDLRTWLSAVLVYPDTCIE 492
Query: 191 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLL 250
G V+ ++ G HV + +N L + D +A +NK+R
Sbjct: 493 GLE----GSIVKGLISSGLDHVMSLVANLLGEVVSGNDDQLA----------TNKDR--- 621
Query: 251 EEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIK 310
+P WI D +LLQ + V AD VVAADGSG++ V +AV+AAP S KRYVI
Sbjct: 622 -------FPSWIRDEDTKLLQANGVTADAVVAADGSGDYAKVMDAVSAAPESSMKRYVIY 780
Query: 311 IKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSAT 358
+K GVY ENVE+ KKK NIM +G+G TII+GSRN VDGSTTF SAT
Sbjct: 781 VKKGVYVENVEIKKKKWNIMLIGEGMDATIISGSRNYVDGSTTFRSAT 924
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 129 bits (325), Expect = 2e-30
Identities = 59/98 (60%), Positives = 70/98 (71%)
Frame = +2
Query: 492 YLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTW 551
YLGRPWK YSRT+FMQS +SD I P GW EWNGNFALNTL Y EY N+GPGAG + RV W
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 552 KGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
G+ V+ ++EA FT FI G+ L STG ++ GL
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSGL 295
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 128 bits (321), Expect = 7e-30
Identities = 57/99 (57%), Positives = 70/99 (70%)
Frame = +1
Query: 491 TYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVT 550
TYLGRPWKEYSRTV+MQS + I+P GW EWNG+FAL+TL Y EY N G G+ T+ RVT
Sbjct: 19 TYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNGDFALSTLYYAEYDNRGAGSSTANRVT 198
Query: 551 WKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
W G+ VI A +A +FT NF+ G W+ TG P+ GL
Sbjct: 199 WPGYHVI-GATDAANFTVSNFVSGDDWIPQTGVPYMSGL 312
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,313,050
Number of Sequences: 36976
Number of extensions: 236783
Number of successful extensions: 1577
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1493
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1511
length of query: 589
length of database: 9,014,727
effective HSP length: 101
effective length of query: 488
effective length of database: 5,280,151
effective search space: 2576713688
effective search space used: 2576713688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137821.5


