

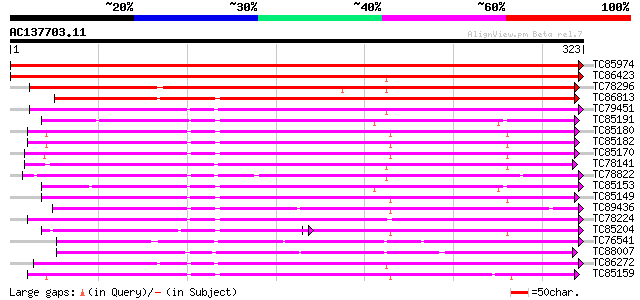
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137703.11 + phase: 0
(323 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85974 PIR|JC4782|JC4782 peroxidase (EC 1.11.1.7) 2 precursor -... 652 0.0
TC86423 homologue to PIR|T09667|T09667 peroxidase (EC 1.11.1.7) ... 484 e-137
TC78296 similar to PIR|T14077|T14077 peroxidase (EC 1.11.1.7) AT... 409 e-115
TC86813 similar to GP|1546694|emb|CAA67338.1 peroxidase ATP20a {... 365 e-101
TC79451 weakly similar to GP|4760700|dbj|BAA77387.1 peroxidase 1... 247 4e-66
TC85191 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B... 237 5e-63
TC85180 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) ... 236 7e-63
TC85182 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) ... 234 4e-62
TC85170 homologue to PIR|JC4781|JC4781 peroxidase (EC 1.11.1.7) ... 231 3e-61
TC78141 similar to GP|5002348|gb|AAD37430.1| peroxidase 5 precur... 228 3e-60
TC78822 similar to PIR|T02962|T02962 peroxidase (EC 1.11.1.7) is... 228 3e-60
TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precurso... 225 2e-59
TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B... 224 3e-59
TC89436 similar to PIR|T09240|T09240 peroxidase (EC 1.11.1.7) pr... 224 3e-59
TC78224 similar to GP|5453379|gb|AAD43561.1| bacterial-induced p... 220 7e-58
TC85204 homologue to GP|13992526|emb|CAC38073. peroxidase1A {Med... 139 8e-58
TC76541 similar to GP|5002238|gb|AAD37376.1| peroxidase {Glycine... 219 1e-57
TC88007 similar to PIR|T05993|T05993 probable peroxidase (EC 1.1... 218 2e-57
TC86272 similar to GP|4204761|gb|AAD11482.1| peroxidase precurso... 217 4e-57
TC85159 homologue to PIR|T09665|T09665 peroxidase (EC 1.11.1.7) ... 217 6e-57
>TC85974 PIR|JC4782|JC4782 peroxidase (EC 1.11.1.7) 2 precursor - alfalfa,
complete
Length = 1258
Score = 652 bits (1683), Expect = 0.0
Identities = 323/323 (100%), Positives = 323/323 (100%)
Frame = +2
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA
Sbjct: 50 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 229
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR
Sbjct: 230 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 409
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF
Sbjct: 410 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 589
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 240
ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA
Sbjct: 590 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 769
Query: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 300
INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM
Sbjct: 770 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 949
Query: 301 TKLGRVGVKNARNGKIRTDCSVL 323
TKLGRVGVKNARNGKIRTDCSVL
Sbjct: 950 TKLGRVGVKNARNGKIRTDCSVL 1018
>TC86423 homologue to PIR|T09667|T09667 peroxidase (EC 1.11.1.7) pxdD
precursor - alfalfa (fragment), complete
Length = 1192
Score = 484 bits (1247), Expect = e-137
Identities = 234/328 (71%), Positives = 282/328 (85%), Gaps = 5/328 (1%)
Frame = +1
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M + N+ILV L LTL L T AQLS +HY N CPNV++IVR AV+KKF QTF TVPA
Sbjct: 46 MAQLNLILVSLLFLTLFLHSRPTHAQLSRHHYKNSCPNVENIVREAVKKKFHQTFTTVPA 225
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCD S+LV+S+ +N+AE+DHP+NLSLAGDGFDTVI+AKAA+DAVP C+
Sbjct: 226 TLRLFFHDCFVQGCDGSILVSSTPHNRAERDHPDNLSLAGDGFDTVIQAKAAVDAVPLCQ 405
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILA+ATRDVI LAGGP Y VELGRFDGL S+ SDVNG+LP+P FNLNQLNTLF
Sbjct: 406 NKVSCADILAMATRDVIALAGGPYYEVELGRFDGLRSKDSDVNGKLPEPGFNLNQLNTLF 585
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNV 235
++GLTQT+MIALSGAHT+GFSHC++F+NR+ + VDPTL+ +YAAQL+ MCPRNV
Sbjct: 586 KHHGLTQTEMIALSGAHTVGFSHCNKFTNRVYNFKTTSRVDPTLDLKYAAQLKSMCPRNV 765
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPR+A++MDP TP FDNVY+KNLQ+GKGLFTSDQ+LFTD+RS+ VN+FA++ +F+AN
Sbjct: 766 DPRVAVDMDPVTPHAFDNVYFKNLQKGKGLFTSDQVLFTDSRSKAAVNAFASSNKIFHAN 945
Query: 296 FITAMTKLGRVGVKNARNGKIRTDCSVL 323
F+ AMTKLGRVGVKN+ NG IRTDCSV+
Sbjct: 946 FVAAMTKLGRVGVKNSHNGNIRTDCSVI 1029
>TC78296 similar to PIR|T14077|T14077 peroxidase (EC 1.11.1.7) ATP8a -
Arabidopsis thaliana, partial (95%)
Length = 1443
Score = 409 bits (1052), Expect = e-115
Identities = 212/351 (60%), Positives = 255/351 (72%), Gaps = 41/351 (11%)
Frame = +1
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
L+L L L T+ AQL+ Y N+CPNV+ +VRSAV +KFQQTFVT PATLRLFFHDCFV
Sbjct: 67 LSLLLLLTATTSSAQLTRGFYNNVCPNVEQLVRSAVNQKFQQTFVTAPATLRLFFHDCFV 246
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
+GCDAS+L+A+ KAE++HP+++SLAGDGFDTV+KAKAA+D P+CRNKVSCADILAL
Sbjct: 247 RGCDASILLATP---KAEREHPDDISLAGDGFDTVVKAKAAVDRDPKCRNKVSCADILAL 417
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT----- 186
ATRDV+NLAGGP Y VELGR DG VS + V LP P FNLNQLN +F +GL+
Sbjct: 418 ATRDVVNLAGGPFYNVELGRRDGRVSTIASVQRSLPGPHFNLNQLNNMFNLHGLSQTDMV 597
Query: 187 -------------------------------QTDMIALSGAHTLGFSHCDRFSNRI---- 211
QTDM+ALSGAHT+GFSHC+RFSNRI
Sbjct: 598 ALSGAHTIGFPHFNCESAIIYIQLNIQMTSHQTDMVALSGAHTIGFSHCNRFSNRIYGFS 777
Query: 212 -QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQ 270
++ +DP+LN QYA QL+QMCP VDPRIAINMDP +P+ FDN Y+KNLQQGKGLFTSDQ
Sbjct: 778 PRSRIDPSLNLQYAFQLRQMCPIRVDPRIAINMDPVSPQKFDNQYFKNLQQGKGLFTSDQ 957
Query: 271 ILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCS 321
+LFTD+RS+ TVN FA+N F + FI A+TKLGRVGVK G+IR DC+
Sbjct: 958 VLFTDSRSKATVNLFASNPKAFESAFINAITKLGRVGVKTGNQGEIRFDCT 1110
>TC86813 similar to GP|1546694|emb|CAA67338.1 peroxidase ATP20a {Arabidopsis
thaliana}, partial (75%)
Length = 1181
Score = 365 bits (936), Expect = e-101
Identities = 185/297 (62%), Positives = 225/297 (75%), Gaps = 1/297 (0%)
Frame = +3
Query: 26 QLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGN 85
QL N Y + CPNV+ +V AV KF QT T ATLRLF HDCFV+GCDASV++AS N
Sbjct: 111 QLVENFYVSSCPNVELVVAQAVTNKFTQTITTGQATLRLFLHDCFVEGCDASVMIASP-N 287
Query: 86 NKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSY 145
AEKD ENLSL GDGFDTVIKAK A+++V C VSCADILA+ATRDVI L GGPS+
Sbjct: 288 GDAEKDAKENLSLPGDGFDTVIKAKQAVESV--CPGVVSCADILAIATRDVIALLGGPSF 461
Query: 146 TVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCD 205
+VELGR DGL S++S+V LP+P+FNLNQLNT+F+ +GL++ DMIALSGAHT+GFSHCD
Sbjct: 462 SVELGRRDGLNSKASNVEANLPKPTFNLNQLNTIFSKHGLSEKDMIALSGAHTVGFSHCD 641
Query: 206 RFSNRI-QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKG 264
+F+NR+ + VDPTL+ YA QL CPRNVDP I + +D T TFDN+YYKNL GKG
Sbjct: 642 QFTNRLYSSQVDPTLDPTYAQQLMSGCPRNVDPNIVLALDTQTEHTFDNLYYKNLVNGKG 821
Query: 265 LFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCS 321
L +SDQ+LFTD SR+TV FA +G+ F F+ A+ KLGRVGVK + G+IR DCS
Sbjct: 822 LLSSDQVLFTDDASRSTVVEFANDGSKFFEAFVVAIKKLGRVGVKTGKEGEIRRDCS 992
>TC79451 weakly similar to GP|4760700|dbj|BAA77387.1 peroxidase 1
{Scutellaria baicalensis}, partial (93%)
Length = 1229
Score = 247 bits (631), Expect = 4e-66
Identities = 136/318 (42%), Positives = 191/318 (59%), Gaps = 6/318 (1%)
Frame = +2
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
+ L + + +QL Y C + IV+ V+K F + +R+ FHDCF+
Sbjct: 74 IVLVIYFLNGNAHSQLEVGFYTYSCGMAEFIVKDEVRKSFNKNPGIAAGLVRMHFHDCFI 253
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
+GCDASVL+ S+ +N AEKD P N GF+ + AKA L+ +C+ VSCADI+A
Sbjct: 254 RGCDASVLLDSTLSNIAEKDSPANKPSLR-GFEVIDNAKAKLEE--ECKGIVSCADIVAF 424
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
A RD + LAGG Y V GR DG +S +SD LP P+FN+NQL LFA GLTQ +M+
Sbjct: 425 AARDSVELAGGLGYDVPAGRRDGKISLASDTRTELPPPTFNVNQLTQLFAKKGLTQDEMV 604
Query: 192 ALSGAHTLGFSHCDRFSNRI----QTPV-DPTLNKQYAAQLQQMCPR-NVDPRIAINMDP 245
LSGAHT+G SHC FS R+ T + DP+L+ YAA L++ CP+ N + + + MDP
Sbjct: 605 TLSGAHTIGRSHCSAFSKRLYNFSSTSIQDPSLDPSYAALLKRQCPQGNTNQNLVVPMDP 784
Query: 246 TTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGR 305
++P T D YY ++ +GLFTSDQ L T+T + V+ A N +++ F AM K+G+
Sbjct: 785 SSPGTADVGYYNDILANRGLFTSDQTLLTNTGTARKVHQNARNPYLWSNKFADAMVKMGQ 964
Query: 306 VGVKNARNGKIRTDCSVL 323
VGV G+IRT+C V+
Sbjct: 965 VGVLTGNAGEIRTNCRVV 1018
>TC85191 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B precursor
- alfalfa, partial (96%)
Length = 1307
Score = 237 bits (604), Expect = 5e-63
Identities = 143/313 (45%), Positives = 187/313 (59%), Gaps = 10/313 (3%)
Frame = +2
Query: 19 IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATL-RLFFHDCFVQGCDAS 77
+P+++ AQLSP+ YA CP +QSIV ++K +T +PA++ RL FHDCFVQGCDAS
Sbjct: 143 LPFSSNAQLSPDFYAKTCPQLQSIVFQILEK-VSKTDSRMPASIIRLHFHDCFVQGCDAS 319
Query: 78 VLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVI 137
VL+ + +E+D N++ D + + K ++ V C NKVSCADIL LA
Sbjct: 320 VLLNKTSTIASEQDAGPNINSLRR-LDVINQIKTEVEKV--CPNKVSCADILTLAAGVSS 490
Query: 138 NLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAH 197
L+GGP + V LGR D L + S N LP PS +L+QL + FA GL D++ALSGAH
Sbjct: 491 VLSGGPGWIVPLGRRDSLTANQSLANRNLPGPSSSLDQLKSSFAAQGLNTVDLVALSGAH 670
Query: 198 TLGFSHC----DRFSNRIQT-PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFD 252
TLG + C DR + T DPTL+ Y QLQ+ CP+N +N DPTTP FD
Sbjct: 671 TLGRARCLFILDRLYDFDNTGKPDPTLDPTYLKQLQKQCPQNGPGNNVVNFDPTTPDKFD 850
Query: 253 NVYYKNLQQGKGLFTSDQILFT----DTRSRNTVNSFATNGNVFNANFITAMTKLGRVGV 308
YY NLQ KGL SDQ LF+ DT S VN+F N NVF NFI +M K+G +GV
Sbjct: 851 KNYYNNLQGKKGLLQSDQELFSTPGADTIS--IVNNFGNNQNVFFQNFINSMIKMGNIGV 1024
Query: 309 KNARNGKIRTDCS 321
+ G+IR C+
Sbjct: 1025LTGKKGEIRKQCN 1063
>TC85180 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B
precursor - alfalfa, complete
Length = 1342
Score = 236 bits (603), Expect = 7e-63
Identities = 139/326 (42%), Positives = 187/326 (56%), Gaps = 15/326 (4%)
Frame = +1
Query: 11 SLALTLCLI-------PYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLR 63
++A+ LC I P+ + AQL+P+ Y+ CPNV SIVR ++ + + + +R
Sbjct: 28 AVAIALCFIVALFGVLPFPSNAQLNPSFYSKTCPNVSSIVREVIRNVSKTDTRMLASLVR 207
Query: 64 LFFHDCFVQGCDASVLVASSGNNKAEKD-HPENLSLAGDGFDTVIKAKAALDAVPQCRNK 122
L FHDCFVQGCDASVL+ ++ +E+D P SL G D V + K A++ C N
Sbjct: 208 LHFHDCFVQGCDASVLLNNTATIVSEQDAFPNRNSLRG--LDVVNQIKTAVEKA--CPNT 375
Query: 123 VSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFAN 182
VSCADILALA L+ GP + V LGR DGL + S N LP P +L+QL FA+
Sbjct: 376 VSCADILALAAELSSTLSQGPDWKVPLGRRDGLTANQSLANQNLPAPFNSLDQLKAAFAS 555
Query: 183 NGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDP 237
GL+ TD++ALSGAHT G +HC F +R+ DPTLN Y QL+ +CP
Sbjct: 556 QGLSTTDLVALSGAHTFGRAHCSLFVSRLYNFSNTGSPDPTLNATYLQQLRNICPNGGPG 735
Query: 238 RIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSR--NTVNSFATNGNVFNAN 295
+ DPTTP FD YY NLQ KGL SDQ LF+ + + + VN+FAT+ F +
Sbjct: 736 TPLASFDPTTPDKFDKNYYSNLQVKKGLLQSDQELFSTSGADTISIVNNFATDQKAFFES 915
Query: 296 FITAMTKLGRVGVKNARNGKIRTDCS 321
F AM K+G +GV G+IR C+
Sbjct: 916 FKAAMIKMGNIGVLTGNQGEIRKQCN 993
>TC85182 homologue to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B
precursor - alfalfa, complete
Length = 1373
Score = 234 bits (596), Expect = 4e-62
Identities = 140/326 (42%), Positives = 182/326 (54%), Gaps = 15/326 (4%)
Frame = +3
Query: 11 SLALTLCLI-------PYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLR 63
++A+ LC I P+++ AQL P+ Y N CPNV SIVR ++ ++ + + +R
Sbjct: 159 AVAIALCCIVVVLGGLPFSSNAQLDPSFYRNTCPNVSSIVREVIRSVSKKDPRMLGSLVR 338
Query: 64 LFFHDCFVQGCDASVLVASSGNNKAEKD-HPENLSLAGDGFDTVIKAKAALDAVPQCRNK 122
L FHDCFVQGCDASVL+ + +E+D P SL G D V + K A++ C N
Sbjct: 339 LHFHDCFVQGCDASVLLNKTDTVVSEQDAFPNRNSLRG--LDVVNQIKTAVEKA--CPNT 506
Query: 123 VSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFAN 182
VSCADILAL+ LA GP + V LGR DGL + N LP P +QL FA
Sbjct: 507 VSCADILALSAELSSTLADGPDWKVPLGRRDGLTANQLLANKNLPAPFNTTDQLKAAFAA 686
Query: 183 NGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDP 237
GL TD++ALSGAHT G +HC F +R+ DPTLN Y QL+ +CP
Sbjct: 687 QGLDTTDLVALSGAHTFGRAHCSLFVSRLYNFNGTGSPDPTLNTTYLQQLRTICPNGGPG 866
Query: 238 RIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSR--NTVNSFATNGNVFNAN 295
N DPTTP FD YY NLQ KGL SDQ LF+ + S + VN FAT+ F +
Sbjct: 867 TNLTNFDPTTPDKFDKNYYSNLQVKKGLLQSDQELFSTSGSDTISIVNKFATDQKAFFES 1046
Query: 296 FITAMTKLGRVGVKNARNGKIRTDCS 321
F AM K+G +GV + G+IR C+
Sbjct: 1047FKAAMIKMGNIGVLTGKQGEIRKQCN 1124
>TC85170 homologue to PIR|JC4781|JC4781 peroxidase (EC 1.11.1.7) 1C
precursor - alfalfa, complete
Length = 1164
Score = 231 bits (589), Expect = 3e-61
Identities = 138/323 (42%), Positives = 183/323 (55%), Gaps = 10/323 (3%)
Frame = +1
Query: 9 VWSLALTLCL--IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
+W + L + L +P+++ AQLSP Y+ CP V SIV + + + + + +RL F
Sbjct: 31 MWCVVLLVVLGGLPFSSDAQLSPTFYSKTCPTVSSIVSNVLTNVSKTDQRMLASLVRLHF 210
Query: 67 HDCFVQGCDASVLVASSGNNKAEKD-HPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSC 125
HDCFV GCDASVL+ ++ +E+ P N SL G D V + K A+++ C N VSC
Sbjct: 211 HDCFVLGCDASVLLNNTATIVSEQQAFPNNNSLRG--LDVVNQIKTAIESA--CPNTVSC 378
Query: 126 ADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGL 185
ADILALA + LA GPS+TV LGR DGL + + N LP P L QL F GL
Sbjct: 379 ADILALAAQASSVLAQGPSWTVPLGRRDGLTANRTLANQNLPAPFNTLVQLKAAFTAQGL 558
Query: 186 TQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIA 240
TD++ALSGAHT G +HC +F R+ DPTLN Y QL+ +CP
Sbjct: 559 NTTDLVALSGAHTFGRAHCAQFVGRLYNFSSTGSPDPTLNTTYLQQLRTICPNGGPGTNL 738
Query: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSR--NTVNSFATNGNVFNANFIT 298
N DPTTP FD YY NLQ KGL SDQ LF+ + + + VN F+T+ N F +F
Sbjct: 739 TNFDPTTPDKFDKNYYSNLQVKKGLLQSDQELFSTSGADTISIVNKFSTDQNAFFESFKA 918
Query: 299 AMTKLGRVGVKNARNGKIRTDCS 321
AM K+G +GV G+IR C+
Sbjct: 919 AMIKMGNIGVLTGTKGEIRKQCN 987
>TC78141 similar to GP|5002348|gb|AAD37430.1| peroxidase 5 precursor
{Phaseolus vulgaris}, partial (92%)
Length = 1254
Score = 228 bits (580), Expect = 3e-60
Identities = 128/319 (40%), Positives = 184/319 (57%), Gaps = 7/319 (2%)
Frame = +2
Query: 9 VWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHD 68
++++ + L L P + AQL+ Y+N CP+V SIVR+ VQ+ Q + RL FHD
Sbjct: 59 IFTVLIFLLLNP--SHAQLTSTFYSNTCPSVSSIVRNVVQQALQNDPRITASLTRLHFHD 232
Query: 69 CFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADI 128
CFV GCDAS+L+ GN + + + + GFD V K K +++ C + VSCADI
Sbjct: 233 CFVNGCDASLLLDQGGNITLSEKNAVPNNNSARGFDVVDKIKTSVEN--SCPSVVSCADI 406
Query: 129 LALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQT 188
LALA ++L+GGPS+ V LGR DGL++ S N +P P+ +L + FA GL +
Sbjct: 407 LALAAEASVSLSGGPSWNVLLGRRDGLIANQSGANTSIPNPTESLANVTAKFAAVGLNTS 586
Query: 189 DMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINM 243
D++ALSGAHT G C F+ R+ DPTLN Y A LQQ CP+N N+
Sbjct: 587 DLVALSGAHTFGRGQCRFFNQRLFNFSGTGKPDPTLNSTYLATLQQNCPQNGSGNTLNNL 766
Query: 244 DPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSR--NTVNSFATNGNVFNANFITAMT 301
DP++P FDN Y+KNL + +GL +DQ LF+ + + VN+FA+N F F+ +M
Sbjct: 767 DPSSPNNFDNNYFKNLLKNQGLLQTDQELFSTNGAATISIVNNFASNQTAFFEAFVQSMI 946
Query: 302 KLGRVGVKNARNGKIRTDC 320
+G + G+IR+DC
Sbjct: 947 NMGNISPLIGSQGEIRSDC 1003
>TC78822 similar to PIR|T02962|T02962 peroxidase (EC 1.11.1.7) isozyme 40K
precursor cationic - common tobacco, partial (70%)
Length = 1181
Score = 228 bits (580), Expect = 3e-60
Identities = 137/321 (42%), Positives = 176/321 (54%), Gaps = 5/321 (1%)
Frame = +2
Query: 8 LVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFH 67
LV S+ + C + + L N Y CP + IV++ + +RL FH
Sbjct: 59 LVASMVI-FCFLGISEGGSLRKNFYKKSCPQAEEIVKNITLQHVSSRPELPAKLIRLHFH 235
Query: 68 DCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCAD 127
DCFV+GCDASVL+ S+ N AEKD NLSLAG FD + K AL+ +C VSCAD
Sbjct: 236 DCFVRGCDASVLLESTAGNTAEKDAIPNLSLAG--FDVIEDIKEALEE--KCPGIVSCAD 403
Query: 128 ILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQ 187
IL LATRD P++ V GR DG VSRS + +P P N+ QL +FAN LT
Sbjct: 404 ILTLATRDAFK--NKPNWEVLTGRRDGTVSRSIEALINIPAPFHNITQLRQIFANKKLTL 577
Query: 188 TDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAIN 242
D++ LSGAHT+G HC+ FSNR+ + DP+LN YA L+ C D +
Sbjct: 578 HDLVVLSGAHTIGVGHCNLFSNRLFNFTGKGDQDPSLNPTYANFLKTKCQGLSDTTTTVE 757
Query: 243 MDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTK 302
MDP + TFDN YY L Q KGLFTSD L T +SRN VN + N F F +M +
Sbjct: 758 MDPNSSTTFDNDYYPVLLQNKGLFTSDAALLTTKQSRNIVNELVSQ-NKFFTEFSQSMKR 934
Query: 303 LGRVGVKNARNGKIRTDCSVL 323
+G + V NG+IR CSV+
Sbjct: 935 MGAIEVLTGSNGEIRRKCSVV 997
>TC85153 similar to GP|2245683|gb|AAC98519.1| peroxidase precursor {Glycine
max}, complete
Length = 1413
Score = 225 bits (573), Expect = 2e-59
Identities = 136/315 (43%), Positives = 183/315 (57%), Gaps = 10/315 (3%)
Frame = +3
Query: 19 IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATL-RLFFHDCFVQGCDAS 77
+P+++ AQL P Y CP + SI V +K +T +PA++ RL FHDCFVQGCDAS
Sbjct: 105 LPFSSNAQLDPYFYGKTCPKLHSIAFK-VLRKVAKTDPRMPASIIRLHFHDCFVQGCDAS 281
Query: 78 VLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVI 137
VL+ ++ +E+D N++ G D + + K ++ C N+VSCADIL LA+
Sbjct: 282 VLLNNTATIVSEQDAFPNINSLR-GLDVINQIKTKVEKA--CPNRVSCADILTLASGISS 452
Query: 138 NLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAH 197
L GGP + V LGR D L + S N LP P+F+L++L + FA GL D++ALSGAH
Sbjct: 453 VLTGGPGWEVPLGRRDSLTANQSLANQNLPGPNFSLDRLKSAFAAQGLNTVDLVALSGAH 632
Query: 198 TLGFSHC----DRFSNRIQT-PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFD 252
T G + C DR N T DPTL+ Y QL+ CP+N +N DPTTP T D
Sbjct: 633 TFGRARCLFILDRLYNFNNTGKPDPTLDTTYLQQLRNQCPQNGTGNNRVNFDPTTPDTLD 812
Query: 253 NVYYKNLQQGKGLFTSDQILFT----DTRSRNTVNSFATNGNVFNANFITAMTKLGRVGV 308
+Y NLQ KGL SDQ LF+ DT S VNSFA + NVF NFI +M K+G + V
Sbjct: 813 KNFYNNLQGKKGLLQSDQELFSTPGADTIS--IVNSFANSQNVFFQNFINSMIKMGNIDV 986
Query: 309 KNARNGKIRTDCSVL 323
+ G+IR C+ +
Sbjct: 987 LTGKKGEIRKQCNFI 1031
>TC85149 similar to PIR|JC4780|JC4780 peroxidase (EC 1.11.1.7) 1B precursor
- alfalfa, partial (95%)
Length = 1274
Score = 224 bits (572), Expect = 3e-59
Identities = 127/310 (40%), Positives = 181/310 (57%), Gaps = 7/310 (2%)
Frame = +2
Query: 19 IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASV 78
+P+++ AQL P+ Y + CP V SI+R ++ + + + +RL FHDCFV GCDASV
Sbjct: 80 LPFSSDAQLDPSFYRDTCPKVHSIIREVIRNVSKTDPRMLASLVRLHFHDCFVLGCDASV 259
Query: 79 LVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVIN 138
L+ + +E++ N++ G D V + K A++ C N VSCADILAL+ +
Sbjct: 260 LLNKTDTIVSEQEAFPNINSLR-GLDVVNQIKTAVEKA--CPNTVSCADILALSAQISSI 430
Query: 139 LAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHT 198
LA GP++ V LGR DGL + S N LP P +L+QL + FA GL+ TD++ALSGAHT
Sbjct: 431 LADGPNWKVPLGRRDGLTANQSLANQNLPAPFNSLDQLKSAFAAQGLSTTDLVALSGAHT 610
Query: 199 LGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDN 253
G + C ++R+ DPTLN Y +L+++CP P N DPTTP FD
Sbjct: 611 FGRARCTFITDRLYNFSSTGKPDPTLNTTYLQELRKICPNGGPPNNLANFDPTTPDKFDK 790
Query: 254 VYYKNLQQGKGLFTSDQILFTDTRSR--NTVNSFATNGNVFNANFITAMTKLGRVGVKNA 311
YY NLQ KGL SDQ LF+ + + + VN F+ + N F +F AM K+G +GV
Sbjct: 791 NYYSNLQGKKGLLQSDQELFSTSGADTISIVNKFSADKNAFFDSFEAAMIKMGNIGVLTG 970
Query: 312 RNGKIRTDCS 321
+ G+IR C+
Sbjct: 971 KKGEIRKHCN 1000
>TC89436 similar to PIR|T09240|T09240 peroxidase (EC 1.11.1.7) prx11
precursor - spinach, partial (91%)
Length = 1127
Score = 224 bits (571), Expect = 3e-59
Identities = 128/304 (42%), Positives = 176/304 (57%), Gaps = 5/304 (1%)
Frame = +1
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
A+L ++Y CP + I+ V LR+FFHDCF++GCDASVL+ S+
Sbjct: 100 AELHAHYYDQTCPQLDKIISETVLTASIHDPKVPARILRMFFHDCFIRGCDASVLLDSTA 279
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
N+AEKD P N+S+ F + +AKA L+ C VSCADILAL RDV+ ++GGP
Sbjct: 280 TNQAEKDGPPNISVRS--FYVIDEAKAKLELA--CPGVVSCADILALLARDVVAMSGGPY 447
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
+ V GR DG VS++SD LP P+ N+ QL FA GL DM+ LSG HTLGFSHC
Sbjct: 448 WKVLKGRKDGRVSKASDT-ANLPAPTLNVGQLIQSFAKRGLGVKDMVTLSGGHTLGFSHC 624
Query: 205 DRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNL 259
F R+ DP LN ++A L+ CP+ + + A +T FDN YYK L
Sbjct: 625 SSFEARLHNFSSVHDTDPRLNTEFALDLKNKCPKPNNNQNAGQFLDSTASVFDNDYYKQL 804
Query: 260 QQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTD 319
GKG+F+SDQ L D R+R V +FA + ++F F +M KLG ++ + NG++R +
Sbjct: 805 LAGKGVFSSDQSLVGDYRTRWIVEAFARDQSLFFKEFAASMLKLG--NLRGSDNGEVRLN 978
Query: 320 CSVL 323
C V+
Sbjct: 979 CRVV 990
>TC78224 similar to GP|5453379|gb|AAD43561.1| bacterial-induced peroxidase
precursor {Gossypium hirsutum}, partial (93%)
Length = 1276
Score = 220 bits (560), Expect = 7e-58
Identities = 126/314 (40%), Positives = 177/314 (56%), Gaps = 1/314 (0%)
Frame = +3
Query: 11 SLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCF 70
+L++ L T AQLSPN YA C N+Q+IVR+ + K Q+ + LRLFFHDCF
Sbjct: 90 TLSIFSLLACSTINAQLSPNFYAKTCSNLQTIVRNEMIKVIQKEARMGASILRLFFHDCF 269
Query: 71 VQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILA 130
V GCDAS+L+ G EK+ N A GF+ + K +++ C+ VSCADILA
Sbjct: 270 VNGCDASILLDDKGTFVGEKNSGPNQGSAR-GFEVIDTIKTSVETA--CKATVSCADILA 440
Query: 131 LATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDM 190
LATRD I L GGPS+ V LGR D + S N ++P PS +L+ L +F N LT D+
Sbjct: 441 LATRDGIALLGGPSWAVPLGRRDARTASQSAANSQIPGPSSDLSTLTRMFQNKSLTLNDL 620
Query: 191 IALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRT 250
LSGAHT+G + C F NRI + +++ A ++ CP + D TP
Sbjct: 621 TVLSGAHTIGQTECQFFRNRIHN--EANIDRNLATLRKRNCPTSGGDTNLAPFDSVTPTK 794
Query: 251 FDNVYYKNLQQGKGLFTSDQILFTDTRSR-NTVNSFATNGNVFNANFITAMTKLGRVGVK 309
FDN YYK+L KGL SDQ+LF S+ + V ++ +G F+ +F AM K+ ++
Sbjct: 795 FDNNYYKDLIANKGLLHSDQVLFNGGGSQISLVRKYSRDGAAFSRDFAAAMVKMSKISPL 974
Query: 310 NARNGKIRTDCSVL 323
NG+IR +C ++
Sbjct: 975 TGTNGEIRKNCRIV 1016
>TC85204 homologue to GP|13992526|emb|CAC38073. peroxidase1A {Medicago
sativa}, complete
Length = 1244
Score = 139 bits (351), Expect(2) = 8e-58
Identities = 75/165 (45%), Positives = 96/165 (57%), Gaps = 7/165 (4%)
Frame = +3
Query: 166 LPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLN 220
LP P+FNL QL + F N LT TD++ALSG HT+G C F +R+ D TLN
Sbjct: 507 LPAPTFNLTQLKSSFDNQNLTTTDLVALSGGHTIGRGQCRFFVDRLYNFSNTGNPDSTLN 686
Query: 221 KQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSR- 279
Y LQ +CP ++DPTTP TFD+ YY NLQ G GLF SDQ LF+ S
Sbjct: 687 TTYLQTLQAICPNGGPGTNLTDLDPTTPDTFDSNYYSNLQVGNGLFQSDQELFSTNGSDT 866
Query: 280 -NTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSVL 323
+ VNSFA N +F NF+ +M K+G +GV G+IRT C+ +
Sbjct: 867 ISIVNSFANNQTLFFENFVASMIKMGHIGVLTGSQGEIRTQCNAV 1001
Score = 102 bits (253), Expect(2) = 8e-58
Identities = 60/153 (39%), Positives = 88/153 (57%)
Frame = +2
Query: 19 IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASV 78
+P++ AQL P+ Y + C NV SIVR + Q + + +RL FHDCFVQGCDAS+
Sbjct: 77 VPFSN-AQLDPSFYNSTCSNVDSIVRGVLTNVSQSDPRMLGSLIRLHFHDCFVQGCDASI 253
Query: 79 LVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVIN 138
L+ + +E+ P N + + G D + + K A++ C N VSCADILAL+ +
Sbjct: 254 LLNDTATIVSEQSAPPN-NNSIRGLDVINQIKTAVENA--CPNTVSCADILALSAEISSD 424
Query: 139 LAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSF 171
LA GP++ V LGR D L + +S + + P F
Sbjct: 425 LANGPTWQVPLGRRDSLTANNSLCSSKSSCPHF 523
>TC76541 similar to GP|5002238|gb|AAD37376.1| peroxidase {Glycine max},
partial (90%)
Length = 1322
Score = 219 bits (557), Expect = 1e-57
Identities = 124/297 (41%), Positives = 169/297 (56%)
Frame = +3
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
LS +A CPN+++IVR + K F+ P LR+FFHDCFVQGCD SVL+
Sbjct: 156 LSYGFFAQTCPNLENIVRKHLTKVFKSDNGQAPGLLRIFFHDCFVQGCDGSVLL---DGK 326
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
E+D P+N + + T+ +A + +C VSCADI LA R+ + L+GGP++
Sbjct: 327 PGERDQPQNGGMRTEALKTIDDIRALVHK--ECGRIVSCADITVLAGREAVFLSGGPNFP 500
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
V LGR DG S S LPQP + +FA TD++ALSGAHT G +HC
Sbjct: 501 VPLGRKDG-TSFSIKGTSNLPQPFNKTDVTLKVFAAQNFDVTDVVALSGAHTFGRAHCGT 677
Query: 207 FSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLF 266
F NR+ +P DPTL+K A L+ CP N + N+D TP TFDN YY +L +GLF
Sbjct: 678 FFNRL-SPADPTLDKTLAQNLKNTCP-NANSGNTANLDIRTPATFDNKYYLDLMNKQGLF 851
Query: 267 TSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSVL 323
TSDQ L D+R++ VN FA N +F F+ A K+ ++ V G+IR C+V+
Sbjct: 852 TSDQDLNIDSRTKGLVNDFAVNQGLFFEKFVNAFIKVSQLNVLVGNQGEIRGKCNVV 1022
>TC88007 similar to PIR|T05993|T05993 probable peroxidase (EC 1.11.1.7)
F17M5.180 - Arabidopsis thaliana, partial (92%)
Length = 1254
Score = 218 bits (555), Expect = 2e-57
Identities = 117/294 (39%), Positives = 173/294 (58%)
Frame = +2
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
L+ N+Y CP V+ +V++ V + A +R+ FHDCF+QGCD S+L+ S+ +N
Sbjct: 152 LNMNYYLMSCPFVEPVVKNIVNRALDNDPTLAAALIRMHFHDCFIQGCDGSILLDSTKDN 331
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
AEKD P NLSL G++ + K L+ +C VSCADILA+A + + AGGP Y
Sbjct: 332 TAEKDSPANLSLR--GYEVIDDIKDELE--NRCPGVVSCADILAMAATEAVFYAGGPVYN 499
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
+ GR DG S+ D LP PSFN ++L T F +G + +M+ALSGAHTLG + C
Sbjct: 500 IPKGRKDGRRSKIEDTR-NLPSPSFNASELITQFGQHGFSAQEMVALSGAHTLGVARCSS 676
Query: 207 FSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLF 266
F NR+ + VDP L+ ++A L + C + + T FDNVY+ L + G+
Sbjct: 677 FKNRL-SQVDPALDTEFARTLSRTCTSGDNAEQPFD---ATRNDFDNVYFNALLRKNGVL 844
Query: 267 TSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDC 320
SDQ L++ R+RN VN++A N +F +F AM K+G + +K NG++R++C
Sbjct: 845 FSDQTLYSSPRTRNIVNAYAMNQAMFFLDFQQAMVKMGLLDIKQGSNGEVRSNC 1006
>TC86272 similar to GP|4204761|gb|AAD11482.1| peroxidase precursor {Glycine
max}, partial (86%)
Length = 1350
Score = 217 bits (553), Expect = 4e-57
Identities = 124/317 (39%), Positives = 181/317 (56%), Gaps = 7/317 (2%)
Frame = +1
Query: 14 LTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQG 73
L LC++ +T AQL Y CP + IV + V + + A +R+ FHDCFV+G
Sbjct: 202 LILCILAASTHAQLELGFYTKSCPKAEQIVANFVHEHIRNAPSLAAALIRMHFHDCFVRG 381
Query: 74 CDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALAT 133
CDASVL+ S+ N +AEK+ P NL++ G FD + + K+ ++A +C VSCADI+AL+
Sbjct: 382 CDASVLLNST-NQQAEKNAPPNLTVRG--FDFIDRIKSLVEA--ECPGVVSCADIIALSA 546
Query: 134 RDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIAL 193
RD I GGP + V GR DG+VS + N +P P N L TLFAN GL D++ L
Sbjct: 547 RDSIAATGGPYWKVPTGRRDGVVSNLLEANQNIPAPFSNFTTLQTLFANQGLDMKDLVLL 726
Query: 194 SGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNV-DPRIAINMDPTT 247
SGAHT+G S C FSNR+ + DP+L+ +YA L+ +N+ D + +DP +
Sbjct: 727 SGAHTIGISLCTSFSNRLYNFTGKGDQDPSLDSEYAKNLKTFKCKNINDNTTIVELDPGS 906
Query: 248 PRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNG-NVFNANFITAMTKLGRV 306
TFD YY + + +GLF SD L T++ ++ V F F A F ++ K+G++
Sbjct: 907 RNTFDLGYYSQVVKRRGLFESDSALLTNSVTKALVTQFLQGSLENFYAEFAKSIEKMGQI 1086
Query: 307 GVKNARNGKIRTDCSVL 323
VK G IR C+++
Sbjct: 1087KVKTGSQGVIRKHCALV 1137
>TC85159 homologue to PIR|T09665|T09665 peroxidase (EC 1.11.1.7) pxdC
precursor - alfalfa, complete
Length = 1340
Score = 217 bits (552), Expect = 6e-57
Identities = 133/326 (40%), Positives = 181/326 (54%), Gaps = 15/326 (4%)
Frame = +2
Query: 11 SLALTLCLI------PYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRL 64
SLA C++ P+++ AQL + Y + CPNV SIVR ++ + + + +RL
Sbjct: 80 SLAALCCVVVVLGGLPFSSNAQLDNSFYRDTCPNVHSIVREVLRNVSKTDPRILASLIRL 259
Query: 65 FFHDCFVQGCDASVLVASSGNNKAEKD-HPENLSLAGDGFDTVIKAKAALDAVPQCRNKV 123
FHDCFVQGCDAS+L+ ++ +E+ N S+ G D V + K A++ C N V
Sbjct: 260 HFHDCFVQGCDASILLNTTSTITSEQTAFGNNNSIRG--LDVVNQIKTAVENA--CPNTV 427
Query: 124 SCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANN 183
SCADILALA LA GP + V LGR D L + + N LP P+FNL QL + F N
Sbjct: 428 SCADILALAAEISSVLANGPDWKVPLGRRDSLTANLTLANINLPSPAFNLTQLKSNFDNQ 607
Query: 184 GLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPR 238
GL TD++ALSGAHT+G C F +R+ DPTLN Y L+ +CP
Sbjct: 608 GLDATDLVALSGAHTIGRGQCRFFVDRLYNFSNTGNPDPTLNTTYLQTLRTICPNGGPGS 787
Query: 239 IAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNT---VNSFATNGNVFNAN 295
++DP TP TFD+ YY NL+ KGLF SDQ+L + T +T VNSF N +F
Sbjct: 788 TLTDLDPATPDTFDSAYYSNLRIQKGLFQSDQVL-SSTSGADTIAIVNSFNNNQTLFFEA 964
Query: 296 FITAMTKLGRVGVKNARNGKIRTDCS 321
F +M K+ R+ V G+IR C+
Sbjct: 965 FKASMIKMSRIKVLTGSQGEIRKQCN 1042
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,259,387
Number of Sequences: 36976
Number of extensions: 115501
Number of successful extensions: 965
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 755
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 769
length of query: 323
length of database: 9,014,727
effective HSP length: 96
effective length of query: 227
effective length of database: 5,465,031
effective search space: 1240562037
effective search space used: 1240562037
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137703.11


