

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137669.5 + phase: 0
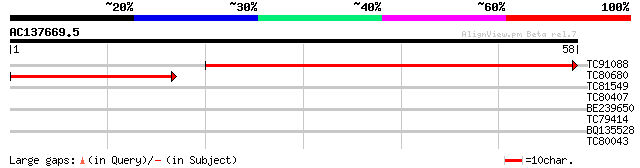
(58 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91088 76 3e-15
TC80680 similar to GP|14517540|gb|AAK62660.1 AT3g50910/F18B3_190... 41 1e-04
TC81549 similar to GP|6630553|gb|AAF19572.1| unknown protein {Ar... 28 0.94
TC80407 similar to GP|10177633|dbj|BAB10781. gene_id:K18B18.4~un... 26 3.6
BE239650 similar to PIR|D87615|D876 conserved hypothetical prote... 25 4.6
TC79414 homologue to GP|12597871|gb|AAG60180.1 unknown protein {... 25 6.1
BQ135528 25 7.9
TC80043 similar to GP|9759561|dbj|BAB11163.1 MtN21 nodulin prote... 25 7.9
>TC91088
Length = 761
Score = 75.9 bits (185), Expect = 3e-15
Identities = 35/38 (92%), Positives = 37/38 (97%)
Frame = -3
Query: 21 TTVIALGTAAVAWSYLPAGGESYSAEDHPVPKHDDAAK 58
TTVIALGTAAVAW+YL AGGESYSA+DHPVPKHDDAAK
Sbjct: 759 TTVIALGTAAVAWAYLRAGGESYSAKDHPVPKHDDAAK 646
>TC80680 similar to GP|14517540|gb|AAK62660.1 AT3g50910/F18B3_190 {Arabidopsis
thaliana}, partial (33%)
Length = 1548
Score = 40.8 bits (94), Expect = 1e-04
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +2
Query: 1 MARRERQRKSKKQKWIW 17
MARRERQRKSKKQKWIW
Sbjct: 1496 MARRERQRKSKKQKWIW 1546
>TC81549 similar to GP|6630553|gb|AAF19572.1| unknown protein {Arabidopsis
thaliana}, partial (7%)
Length = 1309
Score = 27.7 bits (60), Expect = 0.94
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = +2
Query: 27 GTAAVAWSYLPAGGESYSAED 47
G + +AW +LP+ G S+ AED
Sbjct: 1085 GISDLAWQHLPSDGRSFCAED 1147
>TC80407 similar to GP|10177633|dbj|BAB10781. gene_id:K18B18.4~unknown
protein {Arabidopsis thaliana}, partial (7%)
Length = 1159
Score = 25.8 bits (55), Expect = 3.6
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = +3
Query: 27 GTAAVAWSYLPAG 39
GT A AWSY PAG
Sbjct: 846 GTFATAWSYAPAG 884
>BE239650 similar to PIR|D87615|D876 conserved hypothetical protein CC2958
[imported] - Caulobacter crescentus, partial (10%)
Length = 628
Score = 25.4 bits (54), Expect = 4.6
Identities = 9/23 (39%), Positives = 13/23 (56%)
Frame = +3
Query: 31 VAWSYLPAGGESYSAEDHPVPKH 53
+ W L ES++ EDH +P H
Sbjct: 546 IEWEKLNGARESHAQEDHLIPLH 614
>TC79414 homologue to GP|12597871|gb|AAG60180.1 unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 1700
Score = 25.0 bits (53), Expect = 6.1
Identities = 7/13 (53%), Positives = 12/13 (91%)
Frame = +1
Query: 6 RQRKSKKQKWIWG 18
R+++ +K+KWIWG
Sbjct: 457 REQQQQKKKWIWG 495
>BQ135528
Length = 834
Score = 24.6 bits (52), Expect = 7.9
Identities = 8/30 (26%), Positives = 15/30 (49%)
Frame = +2
Query: 6 RQRKSKKQKWIWGSLTTVIALGTAAVAWSY 35
R++ + +QKW+W V +A +Y
Sbjct: 629 REKNTNRQKWLWNLTHCVYLFSNGNIAKTY 718
>TC80043 similar to GP|9759561|dbj|BAB11163.1 MtN21 nodulin protein-like
{Arabidopsis thaliana}, partial (28%)
Length = 1421
Score = 24.6 bits (52), Expect = 7.9
Identities = 11/32 (34%), Positives = 16/32 (49%)
Frame = +1
Query: 13 QKWIWGSLTTVIALGTAAVAWSYLPAGGESYS 44
+ W+ GS+T V+ G WS + G YS
Sbjct: 709 KNWLSGSITKVVRNGRDTSFWSEKWSRGRQYS 804
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.126 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,840,294
Number of Sequences: 36976
Number of extensions: 16849
Number of successful extensions: 92
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 92
length of query: 58
length of database: 9,014,727
effective HSP length: 34
effective length of query: 24
effective length of database: 7,757,543
effective search space: 186181032
effective search space used: 186181032
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 51 (24.3 bits)
Medicago: description of AC137669.5


