

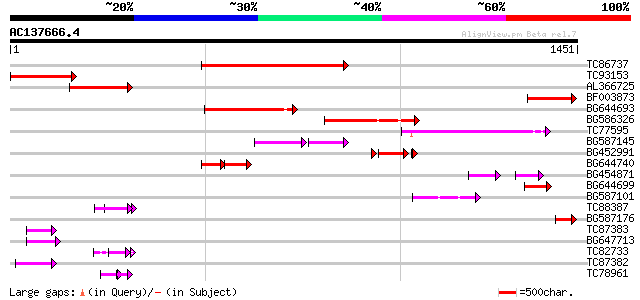
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137666.4 + phase: 0
(1451 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 301 2e-81
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 276 3e-74
AL366725 252 6e-67
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 229 6e-60
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 224 1e-58
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 207 3e-53
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 171 2e-42
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 93 9e-31
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 109 7e-27
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 88 3e-26
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 70 2e-15
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 62 1e-09
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 60 8e-09
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 57 4e-08
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 56 1e-07
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 49 1e-05
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 49 2e-05
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 48 2e-05
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 48 3e-05
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 46 9e-05
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 301 bits (770), Expect = 2e-81
Identities = 168/390 (43%), Positives = 240/390 (61%), Gaps = 14/390 (3%)
Frame = +1
Query: 490 VVNEFHEVF-PDEIPDVPPEREV-EFSIDLVPGAK-----LVSMAPYHMSASELAELKKQ 542
V+ EF ++F P++ VP R + + +I L+P L Y MS EL LKK
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 543 LEDLLDKKFVRPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQ 602
LEDLLDK F++ S S GAPVL V+K G +R C+DYR LN +T K+RYPLP I + + +
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 603 LVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNR 662
+ GA+ F+K+D+ + +H++++KDED +KTAFRTRYG +E+ V PFG+T AP F Y+N+
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINK 882
Query: 663 IFHAFLDKFVVVFIDDILIYSK-IEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSF 721
H FLD FV +IDD+LIY+ +++H ++ VL+ L + L KCEF ++ V +
Sbjct: 883 TLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKY 1062
Query: 722 LGHV-TSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQL 780
+G + T+G G++ DP K+ A+ W P SV RSFLG YY+ FI G+S++ PLT+L
Sbjct: 1063VGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRL 1242
Query: 781 TYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQD--- 837
T K F W A+ E++F +LK+ P+L + PE V D S F LGGVL Q+
Sbjct: 1243TRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGT 1422
Query: 838 --GKVVAYASRQLRVHEKNSPTHDLELAAV 865
VA+ S++L E N P HD EL AV
Sbjct: 1423GAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 276 bits (707), Expect = 3e-74
Identities = 136/171 (79%), Positives = 144/171 (83%), Gaps = 2/171 (1%)
Frame = +2
Query: 1 MVGRNDAAIAAALEDVAQAVGQQQAAANGE--VRMLETFLRNHPPAFKGRYDPDGAQTWL 58
M G +DAA+ AALE VAQAV Q G RMLETFLRNHPP FKGRY PDGA WL
Sbjct: 2 MTGSSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWL 181
Query: 59 KEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVGLLPVLEQDGAVVTWTVFRREFLNRY 118
KE+ERIFRVMQC E QKV+FGTHMLAEEADDWW+ LLPVLEQD AVVTW +FR+EFL RY
Sbjct: 182 KEIERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRY 361
Query: 119 FPEDVRGKKEIEFLELKQGDMSVTEYAAKFTELAKFYPHYTAETAEFSKCI 169
FPEDVRGKKEIEFLELKQGDMSVTEYAAKF ELA FYPHY+AETAEFSKCI
Sbjct: 362 FPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELATFYPHYSAETAEFSKCI 514
>AL366725
Length = 485
Score = 252 bits (644), Expect = 6e-67
Identities = 111/161 (68%), Positives = 138/161 (84%)
Frame = +2
Query: 153 KFYPHYTAETAEFSKCIKFENGLRADIKRSIGYQKIRIFSELVSSCRIYEEDTKAHYKVM 212
KFYPHY AETAEFSKCIKFENGLR DIKR+IGYQ++R+F +LV++CRIYEEDTKAH KV+
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 213 SEKRNKGQSSRPKPYSAPANKGKQRLNDERPQKGKNAPVDVVCYKCGVKGHKSNACTQDE 272
+E++ KGQ SRPKPYSAPA+KGKQR+ D+R K K+AP ++VC+ G KGHKSN C ++
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEI 361
Query: 273 KKCFRCGQKGHVLAECKRGDIVCFSCGEEGHNGAQCTQPKK 313
KKC RC +KGH++A+CKR DIVCF+C EEGH G+QC QPK+
Sbjct: 362 KKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 229 bits (584), Expect = 6e-60
Identities = 113/125 (90%), Positives = 117/125 (93%)
Frame = +2
Query: 1326 GVGRALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHV 1385
GVGRALKS+KLT +FIGPYQISERVGTVAYRVGLPPHL NLHDVFHVSQLRKYVPDPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 1386 IPRDDVQVRDNLTVETLPLRIDDRKVKSLRGKEIPLVRVVWGGATGESLTWELESKMQES 1445
I DDVQVRDNLTVETLP+RIDDRKVK+LRGKEIPLVRVVW A GESLTWELESKM ES
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 1446 YPELF 1450
YPELF
Sbjct: 362 YPELF 376
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 224 bits (572), Expect = 1e-58
Identities = 122/243 (50%), Positives = 158/243 (64%), Gaps = 5/243 (2%)
Frame = +2
Query: 500 DEIPDVPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPW 559
D + VPPE +++F IDL+P + + Y ++ +L LK QL+DLL+K F++PS+ P
Sbjct: 2 DHLL*VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 560 GAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYH 619
G VL +KKKDG +R+ IDY QLN V IK +YPLP ID+L D L G+K F KIDLR G H
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*H 361
Query: 620 QIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDI 679
Q +V ED+ KTAFR RYGHYE VM FG TN P FME MNR+F +LD V+VF +DI
Sbjct: 362 QHRVIGEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDI 541
Query: 680 LIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEF----WLSEVSFLG-HVTSGNGIAVD 734
LIYSK E EH HL++ L++LK+ + C+ L EV F HV SG G+ VD
Sbjct: 542 LIYSKNENEHENHLRLALKVLKD------IGLCQISYV*ILVEVGFFSLHVISGEGLKVD 703
Query: 735 PSK 737
+
Sbjct: 704 SKR 712
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 207 bits (526), Expect = 3e-53
Identities = 115/246 (46%), Positives = 160/246 (64%), Gaps = 1/246 (0%)
Frame = +2
Query: 805 TTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAA 864
T+APIL+LP+ +VVY DAS GLG VL Q KV+AYASRQLR HE N PTHDLE+AA
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 865 VVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFCLNYHPDKA 924
VVF LKIWR YLYG++ ++ +DHKSLKY+F Q ELN+RQRRW+E + DYD + Y+P KA
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 925 KVVADALSRKTLHMSALMVKEFELLEQFRDLSLVCELSSQSVQLGMLKIN-SDFLGSIRE 983
+VADALSR+ + +SA +E + L+ + L+ + LG+ +N +D IR
Sbjct: 365 NLVADALSRRRVDVSA--EREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRL 538
Query: 984 AQQVDVKFVDLMVVSNQAEESDFKVDEQGVLRFRGRICIPDNEELKKLILEEGHKSNLSI 1043
AQ D + Q + ++++ + G + GRI +P++ LK+ I+ E HKS S+
Sbjct: 539 AQGQDENLQKVA----QNDRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSV 706
Query: 1044 HLGATK 1049
H GA +
Sbjct: 707 HPGAPR 724
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 171 bits (432), Expect = 2e-42
Identities = 124/393 (31%), Positives = 188/393 (47%), Gaps = 14/393 (3%)
Frame = +2
Query: 1004 SDFKVDEQGVLRFRGRICIPDNE-------ELKKLILEEGHKSNLSIHLGATKMYQDLKK 1056
S+ ++D L FRGRI +P ++ EL+ +++E H S + H G + + +
Sbjct: 74 SECQLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSR 253
Query: 1057 LFWWSGLKKDVARFVYACLTCQKSKVEHQRPAGLLTPLDVPEWKWDSISMDFVSSLPNT- 1115
F+W G + V RFV C C + Q G L PL VP +SMDF++SLP T
Sbjct: 254 KFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTR 433
Query: 1116 SRGHDSIWVVVDRLTKSAHFIPINISYPVAQLAEIYIQNIVKLHGVPSSIVSDRDPRFTS 1175
RG +WV+VDRL+KS ++ A A+ ++ + HG+P SIVSDR +
Sbjct: 434 GRGSQYLWVIVDRLSKSVTLEEMDTMEAEA-CAQRFLSCHYRFHGMPQSIVSDRGSNWVG 610
Query: 1176 RFWRSLQDALGSKLKLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFT 1235
RFWR G LS++YHPQTDG +ER Q ++ +LR V W LP ++
Sbjct: 611 RFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLA 790
Query: 1236 YNNSYHSSIGMAPFEALYGRKCKTPLCWFESGESVVLGPE-----LVHETTEKVKMIREK 1290
N ++SSIG PF +G P+ E VV E LV + I+ +
Sbjct: 791 LRNRHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAE 967
Query: 1291 MKASQSRQKSYHDKRRKDLE-FQEGGHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISER 1349
+ A+Q R ++ +KRR + +Q G V+L V+ + K L K Y+++
Sbjct: 968 IVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYKSPRPSKKLDWLHHK----YEVTRF 1135
Query: 1350 VGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDP 1382
V + +P ++ FHV LR+ DP
Sbjct: 1136 VTPHVVELNVP---GTVYPKFHVDLLRRAASDP 1225
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 93.2 bits (230), Expect(2) = 9e-31
Identities = 48/133 (36%), Positives = 76/133 (57%)
Frame = +2
Query: 626 EDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKI 685
+D++KTAF T G Y YKVMPFG+ NA + +NR+F L + V+IDD+L+ S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 686 EEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWE 745
+H HLK + L E + +KC F ++ FLG++ + GI V+P ++ A+
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 746 TPKSVTEIRSFLG 758
+PK+ E++ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 60.5 bits (145), Expect(2) = 9e-31
Identities = 35/106 (33%), Positives = 54/106 (50%), Gaps = 4/106 (3%)
Frame = +3
Query: 765 RFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCD 824
RFI + LP +L K FVWD +CE +F +LKQ LTT P+L P+ + +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 825 ASKFGLGGVLMQDG----KVVAYASRQLRVHEKNSPTHDLELAAVV 866
S + VL+++ K + Y S+++ E PT + AV+
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVI 746
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 109 bits (273), Expect(3) = 7e-27
Identities = 54/78 (69%), Positives = 67/78 (85%)
Frame = +3
Query: 943 VKEFELLEQFRDLSLVCELSSQSVQLGMLKINSDFLGSIREAQQVDVKFVDLMVVSNQAE 1002
++ + LEQFRDLSLVCE+S QSV+LGMLKIN++FL SI+EAQ+VDVK VDLM +NQ E
Sbjct: 51 LESWSCLEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTE 230
Query: 1003 ESDFKVDEQGVLRFRGRI 1020
+ DFKVD+QGVL+FR RI
Sbjct: 231 DGDFKVDDQGVLQFRDRI 284
Score = 25.4 bits (54), Expect(3) = 7e-27
Identities = 9/15 (60%), Positives = 14/15 (93%)
Frame = +2
Query: 1028 LKKLILEEGHKSNLS 1042
+KK+ILEE H+SN++
Sbjct: 284 MKKMILEESHRSNVN 328
Score = 25.4 bits (54), Expect(3) = 7e-27
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +1
Query: 926 VVADALSRKTLHM 938
VVAD LSRKTLH+
Sbjct: 1 VVADVLSRKTLHV 39
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 88.2 bits (217), Expect(2) = 3e-26
Identities = 42/69 (60%), Positives = 52/69 (74%)
Frame = -1
Query: 549 KKFVRPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKV 608
K+F +PS+SP GA +L V+KKDG R+CIDYRQ NKVT KN+YPLPRID+L D++
Sbjct: 274 KRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCY 95
Query: 609 FSKIDLRSG 617
F IDLR G
Sbjct: 94 F*NIDLRLG 68
Score = 50.1 bits (118), Expect(2) = 3e-26
Identities = 28/62 (45%), Positives = 38/62 (61%)
Frame = -2
Query: 490 VVNEFHEVFPDEIPDVPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDK 549
V+ F VFPD P +P ERE+ F IDL+ +L+S P M +EL +LK L+D L+K
Sbjct: 450 VLKGFS*VFPDNFPVIPLEREIFFCIDLLLDTQLISNPP*LMDRTELKKLKI*LKDSLEK 271
Query: 550 KF 551
F
Sbjct: 270 GF 265
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 70.5 bits (171), Expect(2) = 2e-15
Identities = 36/81 (44%), Positives = 45/81 (55%)
Frame = +2
Query: 1175 SRFWRSLQDALGSKLKLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEF 1234
S FW+ L G+ L +SSAYHP +DGQSE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1235 TYNNSYHSSIGMAPFEALYGR 1255
YN SY+ S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 31.6 bits (70), Expect(2) = 2e-15
Identities = 21/73 (28%), Positives = 33/73 (44%), Gaps = 2/73 (2%)
Frame = +1
Query: 1294 SQSRQKSYHDKRRKDLEFQEGGHVFLRVTPMTGVGRALKSRKL--TPKFIGPYQISERVG 1351
+Q K DK+R+ EFQ G HV +++ P AL+ + +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 1352 TVAYRVGLPPHLS 1364
A+ PP+LS
Sbjct: 568 ESAFHCKSPPYLS 606
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 62.4 bits (150), Expect = 1e-09
Identities = 31/71 (43%), Positives = 47/71 (65%), Gaps = 1/71 (1%)
Frame = +2
Query: 1317 VFLRVTPMT-GVGRALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQL 1375
V L+V P G R K KL+ ++IGP+++ +R+G VAY + LPP LS +H VFHVS
Sbjct: 8 VLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMF 187
Query: 1376 RKYVPDPSHVI 1386
++Y D +++I
Sbjct: 188 KRYHGDGNYII 220
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 59.7 bits (143), Expect = 8e-09
Identities = 48/179 (26%), Positives = 83/179 (45%), Gaps = 6/179 (3%)
Frame = +2
Query: 1032 ILEEGHKSNLSIHLGATKMYQDLKKL-FWWSGLKKDVARFVYACLTCQKS---KVEHQRP 1087
IL H SN + H +K +++ FWW + KD F+ C CQ+ ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 1088 AGLLTPLDVPEWKWDSISMDFVSSLPNTSRGHDSIWVVVDRLTKSAHFI--PINISYPVA 1145
+ ++V +D +DF+ P+ S + I V VD ++K I P N + V
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFPS-SYNNKYILVAVDYVSKWVEAIASPTNDATVVV 448
Query: 1146 QLAEIYIQNIVKLHGVPSSIVSDRDPRFTSRFWRSLQDALGSKLKLSSAYHPQTDGQSE 1204
++ + I GVP ++SD F ++ + L G + K+++AYHPQ +S+
Sbjct: 449 KM---FKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 57.4 bits (137), Expect = 4e-08
Identities = 31/103 (30%), Positives = 52/103 (50%), Gaps = 5/103 (4%)
Frame = +1
Query: 216 RNKGQSSRPKPYSAPANKGKQR----LNDERP-QKGKNAPVDVVCYKCGVKGHKSNACTQ 270
R++ S R PY + +G + N +RP + P VC+ C + GH ++ C+
Sbjct: 349 RSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGHYVRECPNVAVCHNCSLPGHIASECST 528
Query: 271 DEKKCFRCGQKGHVLAECKRGDIVCFSCGEEGHNGAQCTQPKK 313
+ C+ C + GH+ + C I C +CG+ GH +CT P+K
Sbjct: 529 -KSLCWNCKEPGHMASSCPNEGI-CHTCGKAGHRARECTVPQK 651
Score = 43.5 bits (101), Expect = 6e-04
Identities = 28/84 (33%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Frame = +1
Query: 243 PQKGKNAPVDV-VCYKCGVKGHKSNACTQDEKKCFRCGQKGHVLAECKRGDIVCFSCGEE 301
PQK P D+ +C C +GH + CT +EK C C + GH+ +C D +C C
Sbjct: 643 PQK---PPGDLRLCNNCYKQGHIAVECT-NEKACNNCRKTGHLARDCP-NDPICNLCNIS 807
Query: 302 GHNGAQCTQPKKVRTGGKVFALTG 325
GH QC + + G +L G
Sbjct: 808 GHVARQCPKSNVIGDRGGGGSLRG 879
Score = 40.0 bits (92), Expect = 0.007
Identities = 20/85 (23%), Positives = 33/85 (38%), Gaps = 23/85 (27%)
Frame = +1
Query: 247 KNAPVDVVCYKCGVKGHKSNACTQD----------------------EKKCFRCGQKGHV 284
++ P D +C C + GH + C + + C C Q GH+
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 285 LAECKRGDI-VCFSCGEEGHNGAQC 308
+C G + +C +CG GH +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 34.3 bits (77), Expect = 0.36
Identities = 18/44 (40%), Positives = 19/44 (42%), Gaps = 1/44 (2%)
Frame = +1
Query: 252 DVVCYKCGVKGHKSNACTQDEKK-CFRCGQKGHVLAECKRGDIV 294
DVVC C GH S C C CG +GH EC G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 55.8 bits (133), Expect = 1e-07
Identities = 25/54 (46%), Positives = 37/54 (68%)
Frame = -1
Query: 1397 LTVETLPLRIDDRKVKSLRGKEIPLVRVVWGGATGESLTWELESKMQESYPELF 1450
L +ET P+RI DR K++R K I +V++VW + E +TWE E++M+ YPE F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 48.9 bits (115), Expect = 1e-05
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 2/80 (2%)
Frame = -2
Query: 43 PAFKGRYDPDGAQTWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVGLLPVLEQDG 102
P F+G PD WL+ +ER+F+ + E QKV+ L + A WW + +++G
Sbjct: 724 PDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREG 545
Query: 103 --AVVTWTVFRREFLNRYFP 120
+ TW R++ +Y P
Sbjct: 544 KSKIKTWEKMRQKLTRKYLP 485
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 48.5 bits (114), Expect = 2e-05
Identities = 29/95 (30%), Positives = 47/95 (48%), Gaps = 7/95 (7%)
Frame = +2
Query: 43 PAFKGRYDPDGAQTWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVGLLPVLEQDG 102
P F G+ PD WL+ +ER+F+ + +E QKV+ L + A WW L +G
Sbjct: 347 PDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEG 526
Query: 103 --AVVTWTVFR----REFLN-RYFPEDVRGKKEIE 130
+ TW R R++L+ Y+ ++ KK I+
Sbjct: 527 KSKIKTWDKMRQKLTRKYLHPHYYQDNFTQKKNIQ 631
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 48.1 bits (113), Expect = 2e-05
Identities = 35/119 (29%), Positives = 53/119 (44%), Gaps = 10/119 (8%)
Frame = +3
Query: 214 EKRNKGQSSRPKPYSAPANKGKQRLNDERPQKGKNAPVDVVCYKCGVKGHKSNACTQD-- 271
+K+NK + +P S P GK+ L + G + C+ C H + CTQ
Sbjct: 180 KKKNKFKRKKPDSNSKPRT-GKRPLRVPGMKPGDS------CFICKGLDHIAKFCTQKAE 338
Query: 272 ---EKKCFRCGQKGHVLAECKRGDI-----VCFSCGEEGHNGAQCTQPKKVRTGGKVFA 322
K C RC ++GH C G ++CG+ GH+ A C P ++ GG +FA
Sbjct: 339 WEKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHP--LQEGGTMFA 509
Score = 43.1 bits (100), Expect = 8e-04
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 12/67 (17%)
Frame = +3
Query: 254 VCYKCGVKGHKSNACT-----QDEKKCFRCGQKGHVLAEC----KRGDIV---CFSCGEE 301
+C +C +GH++ C +D K + CG GH LA C + G + CF C E+
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 302 GHNGAQC 308
GH C
Sbjct: 534 GHLSKNC 554
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 47.8 bits (112), Expect = 3e-05
Identities = 28/109 (25%), Positives = 51/109 (46%), Gaps = 3/109 (2%)
Frame = +2
Query: 15 DVAQAVGQQQAAANGEVRMLE-TFLRNHPPAFKGRYDPDGAQTWLKEVERIFRVMQCSEV 73
DV+ + +++ + R L+ ++ P F+G D WL+ +ER+F + E
Sbjct: 542 DVSYSDSSSSRSSHSQRRQLQMNDIKVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEE 721
Query: 74 QKVRFGTHMLAEEADDWWVGLLPVLEQDG--AVVTWTVFRREFLNRYFP 120
QKV+ L + A WW L +++G + TW R++ +Y P
Sbjct: 722 QKVKIVAAKLKKHALIWWENLKRRRKREGKSKIKTWDKMRQKLTRKYLP 868
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 46.2 bits (108), Expect = 9e-05
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Frame = +2
Query: 233 KGKQRLNDERPQKGKNAPVDVV-CYKCGVKGHKSNACTQDE--KKCFRCGQKGHVLAECK 289
+G + D R G+ P C+ CG+ GH + C + KC+RCG++GH+ CK
Sbjct: 290 RGSRDSRDSREYLGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 469
Score = 43.1 bits (100), Expect = 8e-04
Identities = 20/44 (45%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Frame = +2
Query: 274 KCFRCGQKGHVLAECKRGD--IVCFSCGEEGHNGAQC-TQPKKV 314
+CF CG GH +CK GD C+ CGE GH C PKK+
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKL 487
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,539,167
Number of Sequences: 36976
Number of extensions: 659165
Number of successful extensions: 3097
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 2951
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3059
length of query: 1451
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1343
effective length of database: 5,021,319
effective search space: 6743631417
effective search space used: 6743631417
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137666.4


