

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.8 - phase: 0 /pseudo
(676 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
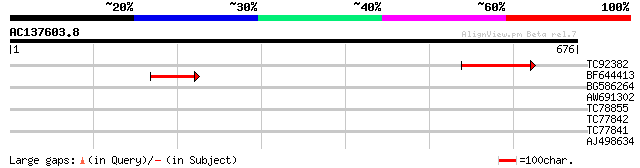
Score E
Sequences producing significant alignments: (bits) Value
TC92382 similar to PIR|E86189|E86189 hypothetical protein [impor... 132 4e-31
BF644413 similar to GP|6642649|gb| putative glucan synthase {Ara... 50 2e-06
BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme n... 30 3.0
AW691302 29 5.2
TC78855 similar to GP|15147869|dbj|BAB62845. Ndr kinase {Arabido... 29 5.2
TC77842 similar to GP|9294383|dbj|BAB02393.1 cytochrome P450 {Ar... 29 5.2
TC77841 weakly similar to GP|9294387|dbj|BAB02397.1 cytochrome P... 29 5.2
AJ498634 similar to GP|20196906|gb expressed protein {Arabidopsi... 29 5.2
>TC92382 similar to PIR|E86189|E86189 hypothetical protein [imported] -
Arabidopsis thaliana, partial (7%)
Length = 739
Score = 132 bits (332), Expect = 4e-31
Identities = 57/88 (64%), Positives = 76/88 (85%)
Frame = +3
Query: 539 AILDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFG 598
A+LD++L+WKA+ MS+H KLRYI K +SGAAWV++L VTYA++W NP GF Q+I++WFG
Sbjct: 285 ALLDIVLSWKARNVMSLHVKLRYIFKAISGAAWVVILPVTYAFSWKNPSGFGQTIKNWFG 464
Query: 599 SNSHSPSMFILAVVVYLSPNMLAAILFL 626
+ S SPS+FILAV +YLSPN+L+AILFL
Sbjct: 465 NGSGSPSIFILAVFIYLSPNILSAILFL 548
Score = 39.3 bits (90), Expect = 0.005
Identities = 18/26 (69%), Positives = 21/26 (80%)
Frame = +3
Query: 513 TAIFNGDVFKKALSVFITAAILKFGQ 538
+ IF+GDVFKK LS+FITAAI K Q
Sbjct: 18 STIFDGDVFKKVLSIFITAAIWKLAQ 95
>BF644413 similar to GP|6642649|gb| putative glucan synthase {Arabidopsis
thaliana}, partial (4%)
Length = 325
Score = 50.4 bits (119), Expect = 2e-06
Identities = 25/58 (43%), Positives = 37/58 (63%)
Frame = +1
Query: 169 SMEVDREILETQDKVAEKTEILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGL 226
S E+ E+ + + TE L+ YNI+P+D ++ AI+ FPE+QAAV AL+ RGL
Sbjct: 103 SEEIPDELKRVMESDSASTEDLIAYNIIPIDATTSTNAIVFFPEVQAAVPALKYFRGL 276
>BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme nc1 -
Arabidopsis thaliana, partial (17%)
Length = 660
Score = 30.0 bits (66), Expect = 3.0
Identities = 15/44 (34%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Frame = -3
Query: 147 LTKAYQTANVLFEVLKAVNMTQSME-VDREILETQDKVAEKTEI 189
LTK YQT N E+++ V ++ + +D E+L+ + V+E +E+
Sbjct: 310 LTKHYQTVNSWKEMIRVVELSHGKKAMDGEVLDRRSFVSEGSEV 179
>AW691302
Length = 1028
Score = 29.3 bits (64), Expect = 5.2
Identities = 14/51 (27%), Positives = 24/51 (46%)
Frame = +1
Query: 67 YLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDA 117
+LC F +A+R P GRG R + A ++ R P ++++ A
Sbjct: 130 FLCCFSGLCRANRAGPPGRGRGSRAVRPASTRKNNRPQRPDYPNKVEEGGA 282
>TC78855 similar to GP|15147869|dbj|BAB62845. Ndr kinase {Arabidopsis
thaliana}, partial (89%)
Length = 1642
Score = 29.3 bits (64), Expect = 5.2
Identities = 11/37 (29%), Positives = 21/37 (56%)
Frame = +3
Query: 105 DPTLKGRIKKSDAREMQSFYQHYYKKYIQALQNAADK 141
D ++ + K A + F +++YK Y+Q LQ+ D+
Sbjct: 198 DVSVSSPVTKQKAAAAKQFIENHYKNYLQGLQDRKDR 308
>TC77842 similar to GP|9294383|dbj|BAB02393.1 cytochrome P450 {Arabidopsis
thaliana}, partial (27%)
Length = 998
Score = 29.3 bits (64), Expect = 5.2
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +3
Query: 152 QTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTE 188
QT N L+ +L T+ E+DREI ++ + + EK E
Sbjct: 630 QTNNPLWGLLATTTKTKMKEIDREIHDSLEGIIEKRE 740
>TC77841 weakly similar to GP|9294387|dbj|BAB02397.1 cytochrome P450
{Arabidopsis thaliana}, partial (38%)
Length = 1299
Score = 29.3 bits (64), Expect = 5.2
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +3
Query: 152 QTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTE 188
QT N L+ +L T+ E+DREI ++ + + EK E
Sbjct: 759 QTNNPLWGLLATTTKTKMKEIDREIHDSLEGIIEKRE 869
>AJ498634 similar to GP|20196906|gb expressed protein {Arabidopsis thaliana},
partial (28%)
Length = 520
Score = 29.3 bits (64), Expect = 5.2
Identities = 16/53 (30%), Positives = 25/53 (46%), Gaps = 9/53 (16%)
Frame = +3
Query: 337 GEAANLRFMPECLCYIYHHMAFEL---------YGMLAGNVSPMTGENIKPAY 380
GE N ++ ECL Y +HH+A ++ Y ++ G S GE+ Y
Sbjct: 252 GEEMNFTYV-ECLLYTFHHLAHKVPNATNSLCGYKIVTGQPSDRLGEDFSEQY 407
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,338,867
Number of Sequences: 36976
Number of extensions: 312987
Number of successful extensions: 1563
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1553
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1562
length of query: 676
length of database: 9,014,727
effective HSP length: 103
effective length of query: 573
effective length of database: 5,206,199
effective search space: 2983152027
effective search space used: 2983152027
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137603.8


