

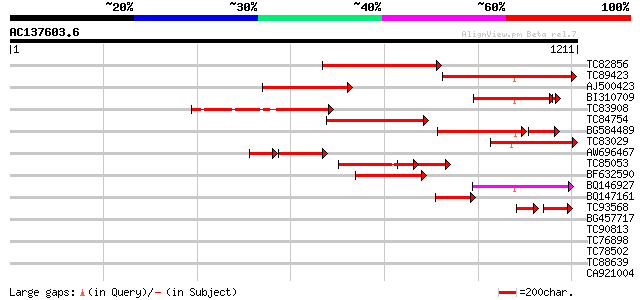
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.6 - phase: 0 /pseudo
(1211 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82856 homologue to PIR|T49914|T49914 callose synthase catalyti... 496 e-140
TC89423 similar to GP|20330757|gb|AAM19120.1 Putative glucan syn... 436 e-122
AJ500423 homologue to PIR|C84727|C8 probable glucan synthase [im... 342 4e-94
BI310709 similar to GP|20330757|g Putative glucan synthase {Oryz... 306 2e-88
TC83908 similar to PIR|T47792|T47792 hypothetical protein F17J16... 319 4e-87
TC84754 homologue to GP|7248390|dbj|BAA92713.1 ESTs AU033035(S15... 307 2e-83
BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.15... 253 7e-80
TC83029 similar to GP|20330757|gb|AAM19120.1 Putative glucan syn... 276 3e-74
AW696467 homologue to PIR|T49914|T4 callose synthase catalytic s... 202 6e-67
TC85053 similar to PIR|E85062|E85062 hypothetical protein AT4g04... 223 3e-58
BF632590 similar to GP|4588012|gb putative callose synthase cata... 178 1e-44
BQ146927 weakly similar to GP|20160606|db putative glucan syntha... 144 3e-34
BQ147161 similar to PIR|T49914|T49 callose synthase catalytic su... 119 5e-27
TC93568 similar to GP|4588012|gb|AAD25952.1| putative callose sy... 84 3e-23
BG457717 similar to PIR|A85045|A85 probable glucan synthase comp... 41 0.002
TC90813 similar to GP|10177205|dbj|BAB10307. cellulose synthase ... 32 1.9
TC76898 similar to GP|21593056|gb|AAM65005.1 GSH-dependent dehyd... 31 2.5
TC78502 similar to GP|8118507|gb|AAF73006.1| NADP-dependent mali... 31 2.5
TC88639 similar to GP|22748334|gb|AAN05336.1 Putative leucine-ri... 31 3.3
CA921004 weakly similar to GP|7110148|gb DNA repair-recombinatio... 30 7.4
>TC82856 homologue to PIR|T49914|T49914 callose synthase catalytic
subunit-like protein - Arabidopsis thaliana, partial
(12%)
Length = 764
Score = 496 bits (1276), Expect = e-140
Identities = 247/255 (96%), Positives = 252/255 (97%)
Frame = +2
Query: 668 TIGQRLLANPLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSEDIFAGFNSTLREGNV 727
TIGQRLLANPLRVRFHYGHPDVFDR+FHLTRGGVSKASKVINLSEDIFAGFNSTLREG+V
Sbjct: 2 TIGQRLLANPLRVRFHYGHPDVFDRLFHLTRGGVSKASKVINLSEDIFAGFNSTLREGSV 181
Query: 728 THHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIG 787
THHEYIQVGKGRDVGLNQ SMFEAKIANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTT+G
Sbjct: 182 THHEYIQVGKGRDVGLNQSSMFEAKIANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTVG 361
Query: 788 FYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLM 847
FYFSTL ITVLTVY+FLYGRLYLVLSGLEEGLS QKAIRDNKPLQVALASQSFVQIGFLM
Sbjct: 362 FYFSTL-ITVLTVYIFLYGRLYLVLSGLEEGLSAQKAIRDNKPLQVALASQSFVQIGFLM 538
Query: 848 ALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGR 907
ALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHY+GRTLLHGGAKYRPTGR
Sbjct: 539 ALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYFGRTLLHGGAKYRPTGR 718
Query: 908 GFVVFHAKFADNYRL 922
GFVVFHAKFADNYRL
Sbjct: 719 GFVVFHAKFADNYRL 763
>TC89423 similar to GP|20330757|gb|AAM19120.1 Putative glucan synthase {Oryza
sativa (japonica cultivar-group)}, partial (17%)
Length = 1020
Score = 436 bits (1120), Expect = e-122
Identities = 206/294 (70%), Positives = 252/294 (85%), Gaps = 9/294 (3%)
Frame = +1
Query: 925 RSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQK 984
RSHFVKGIEL+ILL+VY IFG+ YR ++Y+LIT +WFM GTWL+APFLFNPSGFEWQK
Sbjct: 1 RSHFVKGIELVILLVVYHIFGHAYRGVVAYILITITIWFMAGTWLFAPFLFNPSGFEWQK 180
Query: 985 IVDDWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQY 1044
I+DDWTDW+KWIS RGGIGVPPEKSWESWWE+E EHL++SGMRGI EI+L+LRFFIYQY
Sbjct: 181 ILDDWTDWHKWISNRGGIGVPPEKSWESWWEKEHEHLEHSGMRGIATEIILALRFFIYQY 360
Query: 1045 GLVYHLNFTKSTKSVLPTGL-WHI---MVGDLSNIS-----YLEDFQLVFRLMKGLVFVT 1095
GLVYHL+ T+S +SVL G+ W I ++G + IS DFQLVFRL++G +F+T
Sbjct: 361 GLVYHLSITRSHQSVLVYGISWMIIFLILGLMKGISVGRRRLSADFQLVFRLIEGSIFIT 540
Query: 1096 FVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGY 1155
F++ L+ +IA+A+MT++DI++CILA MPTGWGMLQIAQA KPL+ + G W SV+ LARGY
Sbjct: 541 FLATLIILIAVANMTIKDIIICILAVMPTGWGMLQIAQACKPLIAKTGLWGSVRALARGY 720
Query: 1156 EVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKGRSSR 1209
EVIMGLLLFTP+AFLAWFPFVSEFQTRMLFNQAFS+GLQISRILGGQ++ R+++
Sbjct: 721 EVIMGLLLFTPIAFLAWFPFVSEFQTRMLFNQAFSKGLQISRILGGQKRDRTNK 882
>AJ500423 homologue to PIR|C84727|C8 probable glucan synthase [imported] -
Arabidopsis thaliana, partial (12%)
Length = 581
Score = 342 bits (878), Expect = 4e-94
Identities = 167/193 (86%), Positives = 177/193 (91%)
Frame = -2
Query: 540 KRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKPENQNHAIMFT 599
++I KVYYS L KA + S +E Q LDQVIY+IKLPGPAILGEGKPENQNHAI+FT
Sbjct: 580 RKIDKVYYSALTKAALPTKSIDSSEAVQSLDQVIYRIKLPGPAILGEGKPENQNHAIIFT 401
Query: 600 RGEGLQTIDMNQDNYMEEALKMRNLLQEFLKKHDGVRYPSILGLREHIFTGSVSSLAWFM 659
RGEGLQTIDMNQDNYMEEA KMRNLLQEFLKKH G RYP+ILGLREHIFTGSVSSLAWFM
Sbjct: 400 RGEGLQTIDMNQDNYMEEAFKMRNLLQEFLKKHGGPRYPTILGLREHIFTGSVSSLAWFM 221
Query: 660 SNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSEDIFAGFN 719
SNQETSFVTIGQRLLANPL+VRFHYGHPDVFDR+FHLTRGGVSKASKV+NLSEDIFAGFN
Sbjct: 220 SNQETSFVTIGQRLLANPLKVRFHYGHPDVFDRLFHLTRGGVSKASKVVNLSEDIFAGFN 41
Query: 720 STLREGNVTHHEY 732
STLREGNVTHHEY
Sbjct: 40 STLREGNVTHHEY 2
>BI310709 similar to GP|20330757|g Putative glucan synthase {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 591
Score = 306 bits (784), Expect(2) = 2e-88
Identities = 157/183 (85%), Positives = 163/183 (88%), Gaps = 9/183 (4%)
Frame = +3
Query: 990 TDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYH 1049
TDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYH
Sbjct: 3 TDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYH 182
Query: 1050 LNFTKSTKSVLPTGL-W---HIMVGDLSNIS-----YLEDFQLVFRLMKGLVFVTFVSIL 1100
LNFTKSTKSVL G+ W +++ L +S + DFQLVFRLMKGLVFVTFVSIL
Sbjct: 183 LNFTKSTKSVLVYGISWLVIFLILVILKTVSVGRRKFSADFQLVFRLMKGLVFVTFVSIL 362
Query: 1101 VTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMG 1160
VTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMG
Sbjct: 363 VTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMG 542
Query: 1161 LLL 1163
L
Sbjct: 543 FAL 551
Score = 40.0 bits (92), Expect(2) = 2e-88
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +1
Query: 1160 GLLLFTPVAFLAWFPFV 1176
GLLLFTPVAFLAWFPFV
Sbjct: 541 GLLLFTPVAFLAWFPFV 591
>TC83908 similar to PIR|T47792|T47792 hypothetical protein F17J16.150 -
Arabidopsis thaliana, partial (12%)
Length = 841
Score = 319 bits (818), Expect = 4e-87
Identities = 179/307 (58%), Positives = 215/307 (69%), Gaps = 5/307 (1%)
Frame = +3
Query: 389 IFPDEWNNFLQRVNCSNEEELKEYDELEEELRRWASYRGQTLTRTVRGMMYYRKALELQA 448
I+PDEW NF +R+ N EE + EE +R+WASYRGQTL+RTVRGMMYY +AL LQ
Sbjct: 3 IYPDEWANFDERIKSENFEE-----DREEYVRQWASYRGQTLSRTVRGMMYYWQALLLQY 167
Query: 449 FLDMAKDEDLMEGYKAIENSDDNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRS--- 505
++ A D + EG ++ + ++ + R E Q +A+AD+KF+YVVS Q YG K+S
Sbjct: 168 LIENAGDSGISEGPRSFDYNERDKRLE-----QAKALADLKFTYVVSGQLYGSQKKSKNT 332
Query: 506 -GAARAQDILRLMARYPSLRVAYIDEVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETE 564
+ +IL LM + +LRVAYIDE E+ K KVYYS LVK K
Sbjct: 333 FDRSCYNNILNLMVTHSALRVAYIDETEDT-----KGGKKVYYSVLVKGGEK-------- 473
Query: 565 PEQCLDQVIYKIKLPGPAI-LGEGKPENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRN 623
DQ IY+IKLPGP +GEGKPENQNHAI+FTRGE LQ IDMNQDNY EEA KMRN
Sbjct: 474 ----YDQEIYRIKLPGPPTEIGEGKPENQNHAIIFTRGEALQNIDMNQDNYYEEAFKMRN 641
Query: 624 LLQEFLKKHDGVRYPSILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFH 683
+L+EF H G R P+ILGLREHIFTGSVSSLAWFMSNQETSFVTIGQR+LANPL+VRFH
Sbjct: 642 VLEEF-HAHKGQRKPTILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRVLANPLKVRFH 818
Query: 684 YGHPDVF 690
YGHPD+F
Sbjct: 819 YGHPDIF 839
>TC84754 homologue to GP|7248390|dbj|BAA92713.1 ESTs AU033035(S1515)
D39871(S1515) correspond to a region of the predicted
gene.~Similar to, partial (11%)
Length = 653
Score = 307 bits (786), Expect = 2e-83
Identities = 151/218 (69%), Positives = 175/218 (80%)
Frame = +2
Query: 677 PLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVG 736
PL+VR HYG PDVFDRIFH+TRGG+SKAS+VIN+SEDI+AGFNSTLR GNVTHHEYIQVG
Sbjct: 2 PLKVRMHYGFPDVFDRIFHITRGGISKASRVINISEDIYAGFNSTLRLGNVTHHEYIQVG 181
Query: 737 KGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQIT 796
KGRDVGLNQI++FE K+A GNGEQ LSRD+YRLG FDFFRMLS YFTT+G+Y T+ +T
Sbjct: 182 KGRDVGLNQIALFEGKVAGGNGEQVLSRDIYRLGQLFDFFRMLSFYFTTVGYYVCTM-MT 358
Query: 797 VLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLMALPMLMEIG 856
VLTVY+FLYGR YL SGL+E +S + + N L AL +Q VQIG A+PM+M
Sbjct: 359 VLTVYIFLYGRAYLAFSGLDEAVSEKAKLLGNTALDAALNAQFLVQIGVFTAVPMIMGFI 538
Query: 857 LERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRT 894
LE G A+ FI MQLQL VFFTFSLGTKTHY+GRT
Sbjct: 539 LELGLLKAVFSFITMQLQLCSVFFTFSLGTKTHYFGRT 652
>BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.150 -
Arabidopsis thaliana, partial (12%)
Length = 823
Score = 253 bits (647), Expect(2) = 7e-80
Identities = 112/199 (56%), Positives = 151/199 (75%), Gaps = 9/199 (4%)
Frame = +3
Query: 914 AKFADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPF 973
AKFADNYR+YSRSHFVKG+E++ILLI+Y+++G YRS Y IT MWF+ +WL+APF
Sbjct: 12 AKFADNYRMYSRSHFVKGLEILILLIIYEVYGESYRSSTLYFFITISMWFLAISWLFAPF 191
Query: 974 LFNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEI 1033
LFNPSGF+WQK VDDW+DW +W+ RGGIG+P +KSWESWW+EE EHLKYS +RG I EI
Sbjct: 192 LFNPSGFDWQKTVDDWSDWKRWMGNRGGIGIPSDKSWESWWDEENEHLKYSNVRGKILEI 371
Query: 1034 LLSLRFFIYQYGLVYHLNFTKSTKSVLPTGL-WHIMVGDLSNISYLE--------DFQLV 1084
+L+ RFFIYQYG+VYHLN + +K++L L W ++V L + + DFQL+
Sbjct: 372 VLACRFFIYQYGIVYHLNIARRSKNILVFALSWVVLVIVLIVLKMVSMGRRRFGTDFQLM 551
Query: 1085 FRLMKGLVFVTFVSILVTM 1103
FR++K L+F+ F+S++ +
Sbjct: 552 FRILKALLFLGFLSVMAVL 608
Score = 63.9 bits (154), Expect(2) = 7e-80
Identities = 28/66 (42%), Positives = 41/66 (61%)
Frame = +1
Query: 1109 MTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVA 1168
+T+ D+ +LAFMP+GW ++ IAQ + L W SV+ L+R YE +MGL++F P A
Sbjct: 625 LTVSDLFASVLAFMPSGWAIILIAQTCRGLFEWAKLWASVRRLSRAYEYVMGLIIFMPTA 804
Query: 1169 FLAWFP 1174
L P
Sbjct: 805 VLFVVP 822
>TC83029 similar to GP|20330757|gb|AAM19120.1 Putative glucan synthase {Oryza
sativa (japonica cultivar-group)}, partial (11%)
Length = 692
Score = 276 bits (707), Expect = 3e-74
Identities = 142/194 (73%), Positives = 162/194 (83%), Gaps = 10/194 (5%)
Frame = +2
Query: 1028 GIIAEILLSLRFFIYQYGLVYHLNFTKS-TKSVLPTGLWHIMVG---------DLSNISY 1077
GII EILLSLRFFIYQYGLVYHLN TK +KS L G+ +++ + +
Sbjct: 8 GIIVEILLSLRFFIYQYGLVYHLNITKKGSKSFLVYGISWLVIFVILFVMKTVSVGRRKF 187
Query: 1078 LEDFQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKP 1137
+FQLVFRL+KG++FVTF++ILV +IAL HMT QDI+VCILAFMPTGWGMLQIAQALKP
Sbjct: 188 SANFQLVFRLIKGMIFVTFIAILVILIALPHMTPQDIIVCILAFMPTGWGMLQIAQALKP 367
Query: 1138 LVRRGGFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISR 1197
+VRR GFW SVKTLARGYE++MGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISR
Sbjct: 368 IVRRAGFWGSVKTLARGYEIVMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISR 547
Query: 1198 ILGGQRKGRSSRNK 1211
ILGGQRK R+SR+K
Sbjct: 548 ILGGQRKERASRSK 589
>AW696467 homologue to PIR|T49914|T4 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (6%)
Length = 511
Score = 202 bits (514), Expect(2) = 6e-67
Identities = 98/105 (93%), Positives = 102/105 (96%)
Frame = +1
Query: 575 KIKLPGPAILGEGKPENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEFLKKHDG 634
+IKLPGPAILGEGKPENQNHAI+FTRGEGLQTIDMNQDNYMEEA KMRNLLQEFLKKH G
Sbjct: 178 RIKLPGPAILGEGKPENQNHAIIFTRGEGLQTIDMNQDNYMEEAFKMRNLLQEFLKKHGG 357
Query: 635 VRYPSILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLR 679
RYP+ILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPL+
Sbjct: 358 PRYPTILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLK 492
Score = 72.0 bits (175), Expect(2) = 6e-67
Identities = 36/59 (61%), Positives = 43/59 (72%)
Frame = +3
Query: 513 ILRLMARYPSLRVAYIDEVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQ 571
IL+LM +YPSLRVAYIDEVEEPSK+ ++I KVYYS L KA + S +E Q LDQ
Sbjct: 3 ILKLMTKYPSLRVAYIDEVEEPSKDSSRKIDKVYYSALTKAALPTKSIDSSEAVQSLDQ 179
>TC85053 similar to PIR|E85062|E85062 hypothetical protein AT4g04970
[imported] - Arabidopsis thaliana, partial (14%)
Length = 769
Score = 223 bits (568), Expect = 3e-58
Identities = 112/171 (65%), Positives = 136/171 (79%)
Frame = +1
Query: 702 SKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQT 761
SKAS+VIN+SEDIFAGFN TLR GNVTHHEYIQVGKGRDVGLNQISMFEAK+A+GNGEQ
Sbjct: 1 SKASRVINISEDIFAGFNCTLRGGNVTHHEYIQVGKGRDVGLNQISMFEAKVASGNGEQV 180
Query: 762 LSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLST 821
LSRDVYRLGHR DFFRMLS ++TTIGFYF+++ + V+TVY FL+GRLY+ LSG+E+
Sbjct: 181 LSRDVYRLGHRLDFFRMLSVFYTTIGFYFNSM-VVVMTVYAFLWGRLYMALSGIEK--EA 351
Query: 822 QKAIRDNKPLQVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQ 872
Q +NK L + Q +Q+G ALPM++E LE GF A+ +F+ MQ
Sbjct: 352 QNNASNNKALGAIVNQQFIIQLGIFTALPMVVENTLEHGFLPAVWDFLTMQ 504
Score = 114 bits (284), Expect = 3e-25
Identities = 61/113 (53%), Positives = 74/113 (64%)
Frame = +2
Query: 829 KPLQVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKT 888
KPL + S + F + L++I L G QL +FFTFSLGT+T
Sbjct: 374 KPLVQS*ISSL*SSLVFSLPSQWLLKILLSTGSFLQYGTS*QCSXQLGSLFFTFSLGTRT 553
Query: 889 HYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRLYSRSHFVKGIELMILLIVY 941
H++GRT+LHGGAKYR TGRGFVV H FA+NYRLY+RSHFVK IEL I+LIVY
Sbjct: 554 HFFGRTILHGGAKYRATGRGFVVEHKSFAENYRLYARSHFVKAIELGIILIVY 712
>BF632590 similar to GP|4588012|gb putative callose synthase catalytic
subunit {Gossypium hirsutum}, partial (7%)
Length = 452
Score = 178 bits (452), Expect = 1e-44
Identities = 89/151 (58%), Positives = 111/151 (72%)
Frame = +3
Query: 739 RDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITVL 798
RDVGLNQI++FE K+++GNGEQ LSRD+YRLG FDFFRM+S YFTT+G+YF T+ +TVL
Sbjct: 3 RDVGLNQIALFEGKVSSGNGEQVLSRDIYRLGQLFDFFRMMSFYFTTVGYYFCTM-LTVL 179
Query: 799 TVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLMALPMLMEIGLE 858
TVY FLYG+ YL LSG+ E + + I N L AL +Q QIG A+PM++ LE
Sbjct: 180 TVYAFLYGKTYLALSGVGEIIEERAKITKNTALSAALNTQFLFQIGIFTAVPMVLGFVLE 359
Query: 859 RGFRTALSEFILMQLQLAPVFFTFSLGTKTH 889
+GF A+ FI MQ QL VFFTFSLGT+TH
Sbjct: 360 QGFLRAVVNFITMQFQLCTVFFTFSLGTRTH 452
>BQ146927 weakly similar to GP|20160606|db putative glucan synthase {Oryza
sativa (japonica cultivar-group)}, partial (31%)
Length = 694
Score = 144 bits (362), Expect = 3e-34
Identities = 75/226 (33%), Positives = 118/226 (52%), Gaps = 9/226 (3%)
Frame = +3
Query: 988 DWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLV 1047
D+ D+ WI G + E+SWE WW EEQ+HLK +G+ G + EI+L LRFF +QYG+V
Sbjct: 3 DFDDFMNWIWYSGSVFAKAEQSWERWWYEEQDHLKVTGLWGKLLEIILXLRFFFFQYGIV 182
Query: 1048 YHLNFTKSTKSVLPTGLWHIMVGDLSNI---------SYLEDFQLVFRLMKGLVFVTFVS 1098
Y L + S+ L I V +S I Y + +RL++ LV + +
Sbjct: 183 YQLGISAGNNSIAVYLLSWIYVVVVSGIYAVVVYARNKYSAKEHIYYRLVQFLVIILAIL 362
Query: 1099 ILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVI 1158
++V ++ DI +LAF+PTGWG+L IAQ +P ++ W V AR Y+++
Sbjct: 363 LIVALLEFTEFKFVDIFTSLLAFLPTGWGLLLIAQVFRPFLQSTIIWSGVXAXARLYDIL 542
Query: 1159 MGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRK 1204
G+++ TP A L+ ++S++ F+ S G +G K
Sbjct: 543 FGVIIMTPXALLSXGAWLSKYANLXFFSTKHSXGALPXXXIGTGEK 680
>BQ147161 similar to PIR|T49914|T49 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (4%)
Length = 266
Score = 119 bits (299), Expect = 5e-27
Identities = 56/88 (63%), Positives = 66/88 (74%), Gaps = 1/88 (1%)
Frame = +2
Query: 909 FVVFHAKFADNY-RLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGT 967
FVVFHAKFA L+++S +VY+IF + YRS ++Y+LIT MWFMVGT
Sbjct: 2 FVVFHAKFA**L*TLFAKSLCQGY*ASWYYFVVYEIFXHSYRSAVAYILITVSMWFMVGT 181
Query: 968 WLYAPFLFNPSGFEWQKIVDDWTDWNKW 995
WL+APFLFNPSGFEWQKIVDDWTDWNKW
Sbjct: 182 WLFAPFLFNPSGFEWQKIVDDWTDWNKW 265
>TC93568 similar to GP|4588012|gb|AAD25952.1| putative callose synthase
catalytic subunit {Gossypium hirsutum}, partial (6%)
Length = 579
Score = 84.3 bits (207), Expect(2) = 3e-23
Identities = 37/62 (59%), Positives = 50/62 (79%)
Frame = +3
Query: 1140 RRGGFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRIL 1199
+R W+ +++LAR Y+ MG+L+F P+AF +WFPFVS FQTR++FNQAFSRGL+IS IL
Sbjct: 174 KRLPLWKLIRSLARLYDAGMGMLIFVPIAFFSWFPFVSTFQTRLMFNQAFSRGLEISLIL 353
Query: 1200 GG 1201
G
Sbjct: 354 AG 359
Score = 43.5 bits (101), Expect(2) = 3e-23
Identities = 20/47 (42%), Positives = 31/47 (65%)
Frame = +1
Query: 1083 LVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGML 1129
LV R ++GL + ++ LV I L +++ D+ ILAF+PTGWG+L
Sbjct: 1 LVLRFVQGLSLLLALAGLVVAIILTDLSVPDVFASILAFIPTGWGIL 141
>BG457717 similar to PIR|A85045|A85 probable glucan synthase component
[imported] - Arabidopsis thaliana, partial (1%)
Length = 657
Score = 41.2 bits (95), Expect = 0.002
Identities = 18/24 (75%), Positives = 23/24 (95%)
Frame = +2
Query: 1180 QTRMLFNQAFSRGLQISRILGGQR 1203
QTR+LFN+AFSRGLQISRI+ G++
Sbjct: 8 QTRILFNEAFSRGLQISRIVSGKK 79
>TC90813 similar to GP|10177205|dbj|BAB10307. cellulose synthase catalytic
subunit {Arabidopsis thaliana}, partial (21%)
Length = 925
Score = 31.6 bits (70), Expect = 1.9
Identities = 12/26 (46%), Positives = 14/26 (53%)
Frame = -1
Query: 989 WTDWNKWISIRGGIGVPPEKSWESWW 1014
W WN+WISIR G+ P WW
Sbjct: 298 WLQWNRWISIRHGMN--PVSMIHIWW 227
>TC76898 similar to GP|21593056|gb|AAM65005.1 GSH-dependent dehydroascorbate
reductase 1 putative {Arabidopsis thaliana}, partial
(98%)
Length = 971
Score = 31.2 bits (69), Expect = 2.5
Identities = 19/62 (30%), Positives = 32/62 (50%), Gaps = 6/62 (9%)
Frame = +1
Query: 167 YASFKSIIRYLVQGDREKQVIEYILSEVDKHIE------AGDLISEFKLSALPSLYGQFV 220
+ASF S ++ D +Q + L+ +D+H++ AG+ ++ LS P LY V
Sbjct: 349 FASFSSFLKSKDSNDGTEQALLAELNALDEHLKANGPFVAGEKVTAVDLSLAPKLYHLVV 528
Query: 221 AL 222
AL
Sbjct: 529 AL 534
>TC78502 similar to GP|8118507|gb|AAF73006.1| NADP-dependent malic protein
{Ricinus communis}, partial (65%)
Length = 1540
Score = 31.2 bits (69), Expect = 2.5
Identities = 14/36 (38%), Positives = 19/36 (51%)
Frame = +1
Query: 1110 TLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFW 1145
T ++ ++A + GWG L Q L P RRGG W
Sbjct: 427 TASVVLAGLVAALKLGWGKLS*PQILIPWCRRGGHW 534
>TC88639 similar to GP|22748334|gb|AAN05336.1 Putative leucine-rich repeat
transmembrane protein kinase {Oryza sativa (japonica
cultivar-group)}, partial (28%)
Length = 869
Score = 30.8 bits (68), Expect = 3.3
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +1
Query: 374 SPNEDGVSILFYLQKIFPDEWNNFLQRVNCSNEEELKEYDELEEEL 419
S NE+GV + ++Q I DEWN + + EL Y +EEE+
Sbjct: 358 SLNEEGVDLPRWVQSIVQDEWNTEV------FDMELLRYQSVEEEM 477
>CA921004 weakly similar to GP|7110148|gb DNA repair-recombination protein
{Arabidopsis thaliana}, partial (11%)
Length = 809
Score = 29.6 bits (65), Expect = 7.4
Identities = 15/53 (28%), Positives = 29/53 (54%)
Frame = +1
Query: 146 DRELRKRIEFDNYMSCAVRECYASFKSIIRYLVQGDREKQVIEYILSEVDKHI 198
D++L+K I + A+R+C A F + + D+E V++ + E DK++
Sbjct: 658 DKDLKKGI------TPALRDCVAEFDDLNAKYREADKESNVLQMEVQEADKYL 798
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,287,279
Number of Sequences: 36976
Number of extensions: 519897
Number of successful extensions: 2501
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 2457
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2483
length of query: 1211
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1104
effective length of database: 5,058,295
effective search space: 5584357680
effective search space used: 5584357680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137603.6


