

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.10 + phase: 0
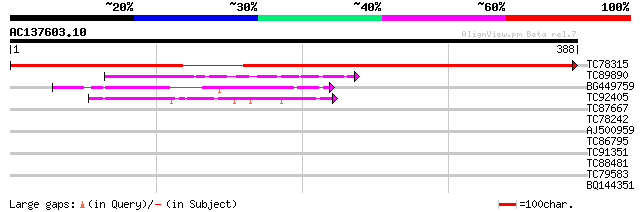
(388 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78315 similar to GP|21593276|gb|AAM65225.1 unknown {Arabidopsi... 668 0.0
TC89890 weakly similar to GP|17065226|gb|AAL32767.1 Unknown prot... 64 8e-11
BG449759 weakly similar to GP|17065226|gb Unknown protein {Arabi... 55 5e-08
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 42 5e-04
TC87667 similar to PIR|T03635|T03635 plastid-lipid-associated pr... 39 0.004
TC78242 similar to GP|17065042|gb|AAL32675.1 Unknown protein {Ar... 37 0.013
AJ500959 similar to GP|17065042|gb Unknown protein {Arabidopsis ... 37 0.017
TC86795 similar to GP|4105180|gb|AAD02288.1| plastoglobule assoc... 35 0.038
TC91351 homologue to GP|16266960|dbj|BAB70118. NADH dehydrogenas... 29 2.7
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 28 4.6
TC79583 similar to GP|18491295|gb|AAL69472.1 At2g42130/T24P15.4 ... 28 6.0
BQ144351 similar to GP|7242771|emb conserved hypothetical protei... 28 6.0
>TC78315 similar to GP|21593276|gb|AAM65225.1 unknown {Arabidopsis
thaliana}, partial (68%)
Length = 1171
Score = 668 bits (1724), Expect = 0.0
Identities = 348/388 (89%), Positives = 348/388 (89%)
Frame = +2
Query: 1 MALTLRVVNVNSIGFESCSRCSTFISPNPSNSRFVSAGCSKVEQISIVTEESENSLIQAL 60
MALTLRVVNVNSIGFESCSRCSTFISPNPSNSRFVSAGCSKVEQISIVTEESENSLIQAL
Sbjct: 11 MALTLRVVNVNSIGFESCSRCSTFISPNPSNSRFVSAGCSKVEQISIVTEESENSLIQAL 190
Query: 61 VGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQR 120
VGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQ
Sbjct: 191 VGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQ- 367
Query: 121 TFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFS 180
AAASIGDGKRILFRFDRAAFS
Sbjct: 368 ---------------------------------------AAASIGDGKRILFRFDRAAFS 430
Query: 181 FKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLL 240
FKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLL
Sbjct: 431 FKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLL 610
Query: 241 TAISSGVGVREAIDKLISLNKNSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQ 300
TAISSGVGVREAIDKLISLNKNSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQ
Sbjct: 611 TAISSGVGVREAIDKLISLNKNSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQ 790
Query: 301 IVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFIL 360
IVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFIL
Sbjct: 791 IVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFIL 970
Query: 361 EILFNDGKVRISRGDNEIIFVHARTNAQ 388
EILFNDGKVRISRGDNEIIFVHARTNAQ
Sbjct: 971 EILFNDGKVRISRGDNEIIFVHARTNAQ 1054
>TC89890 weakly similar to GP|17065226|gb|AAL32767.1 Unknown protein
{Arabidopsis thaliana}, partial (76%)
Length = 1013
Score = 64.3 bits (155), Expect = 8e-11
Identities = 49/174 (28%), Positives = 86/174 (49%)
Frame = +1
Query: 66 RGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGV 125
RG ++P+ +++ + LE + V +P NS L+ G+W+L++TT + F+
Sbjct: 325 RGAEATPEDQQRVDKIARKLEAMNSVKEPLNSDLLNGKWELLYTTSQSILQTQRPKFLRP 504
Query: 126 DFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLP 185
+ ++Q + T+ R NI ++ +A A++ + R A F F
Sbjct: 505 N-GKIYQAI--NTDTLRAQNIETW-----PFYNQATANL-----VPLNSRRVAVKFDF-- 639
Query: 186 FKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
FK+ +P K G +G L+ TYL +LRISRGN+G F+L K +P ++
Sbjct: 640 FKIASLIPIKSSG-SGRGQLEITYLDE--DLRISRGNRGNLFIL-KMVDPSYRV 789
>BG449759 weakly similar to GP|17065226|gb Unknown protein {Arabidopsis
thaliana}, partial (69%)
Length = 634
Score = 55.1 bits (131), Expect = 5e-08
Identities = 51/198 (25%), Positives = 86/198 (42%), Gaps = 5/198 (2%)
Frame = +3
Query: 30 SNSRFVSAGCSKVEQISIVTEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIG 89
+N F A K + S + EE L+ A+ + RG ++P+ ++++ + LE +
Sbjct: 126 TNVSFFPAIFKKGKDASTIKEE----LLDAIASLD-RGADATPEDQQSVDQIARQLEAVN 290
Query: 90 GVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPR----VTN 145
P SSL++G+W+LI+TT + LQT P+ VTN
Sbjct: 291 PTKQPLKSSLLDGKWELIYTT---------------------SQSILQTKRPKLLRSVTN 407
Query: 146 IVSF-SDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGW 204
+ +D + +E+ L + + KF FK+ +P K D A+G
Sbjct: 408 YQAINADTLRAQNMESGPFFNQVTADLTPINAKKVAVKFDTFKIGGLIPVK-APDTARGE 584
Query: 205 LDTTYLSHSGNLRISRGN 222
L+ TYL L +SRG+
Sbjct: 585 LEITYLDE--ELXVSRGD 632
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 41.6 bits (96), Expect = 5e-04
Identities = 52/195 (26%), Positives = 89/195 (44%), Gaps = 25/195 (12%)
Frame = +2
Query: 55 SLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPT-NSSLIEGRWQLIFT---- 109
+L+ A+ G+ RG ++S + L + A + LE GG+ D T N ++GRW+LI++
Sbjct: 215 NLLSAVSGLN-RGLAASEEDLQKADAAAKELEDAGGLVDLTDNLDRLQGRWKLIYSSAFS 391
Query: 110 ------TRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDA----IGELKVE 159
+RPG PI R + + VFQ + + + D NIV + L+V
Sbjct: 392 SRTLGGSRPG--PPIGR-LLPITLGQVFQRIDILSKD--FDNIVDLQLGAPWPLPPLEVT 556
Query: 160 AAAS-----IGDGK-RILFRFDRAAFSFKFL---PFKVP-YPVPFKLLGDEAKGWLDTTY 209
A + +G K +I+F + F P +P P + + G + TY
Sbjct: 557 ATLAHKFELVGSSKIKIIFEKTTVKTTGTFSQLPPLDLPQIPDALRPQSNRGSGDFEVTY 736
Query: 210 LSHSGNLRISRGNKG 224
L + R++RG++G
Sbjct: 737 L--DADTRVTRGDRG 775
>TC87667 similar to PIR|T03635|T03635 plastid-lipid-associated protein pap -
common tobacco (fragment), partial (84%)
Length = 1179
Score = 38.5 bits (88), Expect = 0.004
Identities = 20/42 (47%), Positives = 26/42 (61%)
Frame = +1
Query: 191 PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQ 232
P+ + D A+ WL TTYL LRISRG+ G+ FVL K+
Sbjct: 823 PLKIPISNDNAQSWLLTTYLDEE--LRISRGDGGSVFVLIKE 942
>TC78242 similar to GP|17065042|gb|AAL32675.1 Unknown protein {Arabidopsis
thaliana}, partial (77%)
Length = 1180
Score = 37.0 bits (84), Expect = 0.013
Identities = 34/116 (29%), Positives = 55/116 (47%), Gaps = 3/116 (2%)
Frame = +3
Query: 20 RCSTFISPNPSNSRFVS-AGCSKVEQISIVTEESENSLIQALVGIQG--RGRSSSPQQLN 76
R T + N S+ +S A S V Q E EN+ + L +Q RG ++P Q +
Sbjct: 114 RSHTLNNVNRRKSKSISMAMASSVTQA--YDSELENNKHELLTSVQDTQRGLLTTPHQRS 287
Query: 77 AIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQ 132
+IE A+ +E + DP + + ++G W+L +T+ P Q + FF V Q
Sbjct: 288 SIEEALVNVEG-SNMGDPIDFNKLDGTWRLQYTSAPDVLILFQAA-ATLPFFQVGQ 449
>AJ500959 similar to GP|17065042|gb Unknown protein {Arabidopsis thaliana},
partial (38%)
Length = 717
Score = 36.6 bits (83), Expect = 0.017
Identities = 29/96 (30%), Positives = 49/96 (50%), Gaps = 3/96 (3%)
Frame = +2
Query: 20 RCSTFISPNPSNSRFVS-AGCSKVEQISIVTEESENSLIQALVGIQG--RGRSSSPQQLN 76
R T + N S+ +S A S V Q E EN+ + L +Q RG ++P Q +
Sbjct: 86 RSHTLNNVNRRKSKSISMAMASSVTQA--YDSELENNKHELLTSVQDTQRGLLTTPHQRS 259
Query: 77 AIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRP 112
+IE A+ +E + DP + + ++G W+L +T+ P
Sbjct: 260 SIEEALVNVEG-SNMGDPIDFNKLDGTWRLQYTSAP 364
>TC86795 similar to GP|4105180|gb|AAD02288.1| plastoglobule associated protein
PG1 precursor {Pisum sativum}, partial (95%)
Length = 1400
Score = 35.4 bits (80), Expect = 0.038
Identities = 22/51 (43%), Positives = 28/51 (54%)
Frame = +2
Query: 185 PFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
P K+P P G+ WL TTYL +LRISRG+ G FVL ++ P
Sbjct: 1043 PLKIPIP------GERTSSWLLTTYLDK--DLRISRGD-GGLFVLAREGSP 1168
>TC91351 homologue to GP|16266960|dbj|BAB70118. NADH dehydrogenase subunit 1
{Zu cristatus}, partial (4%)
Length = 982
Score = 29.3 bits (64), Expect = 2.7
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = -3
Query: 206 DTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKNSG 264
DTT SH+ L+ S K + +LQ LL + ++E++ L++L +N G
Sbjct: 854 DTTSQSHAVTLKSSENTKKSLHILQDLFSKDFSLLIHPGRSIQMKESLRYLLNLPQNEG 678
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 28.5 bits (62), Expect = 4.6
Identities = 15/32 (46%), Positives = 18/32 (55%)
Frame = -2
Query: 26 SPNPSNSRFVSAGCSKVEQISIVTEESENSLI 57
SP NSRF A C V+ IS T S +SL+
Sbjct: 947 SPISLNSRFKFAACFNVQSISASTSSSGSSLL 852
>TC79583 similar to GP|18491295|gb|AAL69472.1 At2g42130/T24P15.4
{Arabidopsis thaliana}, partial (51%)
Length = 704
Score = 28.1 bits (61), Expect = 6.0
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +1
Query: 67 GRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRW 104
G+++ Q+++ ER I LE + PT S +EGRW
Sbjct: 382 GKTTEYQRIDVNER-ITGLERLNPTPRPTTSPFLEGRW 492
>BQ144351 similar to GP|7242771|emb conserved hypothetical protein SCD8A.23
{Streptomyces coelicolor A3(2)}, partial (2%)
Length = 1019
Score = 28.1 bits (61), Expect = 6.0
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = +3
Query: 345 GGPFGYPLEFGKKFILEILFNDGKVRISRG 374
GGPF PL K L LFN K++ RG
Sbjct: 573 GGPFWAPLGKKKTLFLNPLFNKKKIKRGRG 662
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,558,598
Number of Sequences: 36976
Number of extensions: 131071
Number of successful extensions: 717
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 714
length of query: 388
length of database: 9,014,727
effective HSP length: 98
effective length of query: 290
effective length of database: 5,391,079
effective search space: 1563412910
effective search space used: 1563412910
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137603.10


