

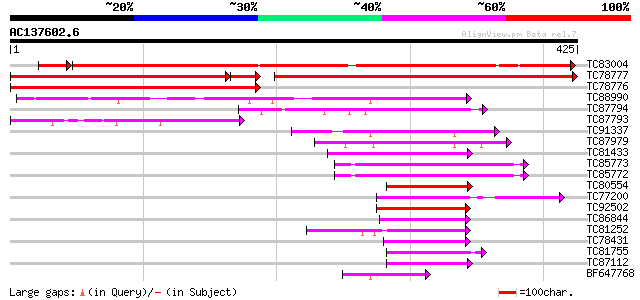
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.6 + phase: 0
(425 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83004 similar to PIR|T10985|T10985 regulator protein ROM2 - ki... 518 e-155
TC78777 similar to PIR|T10985|T10985 regulator protein ROM2 - ki... 437 e-127
TC78776 similar to PIR|T07150|T07150 G-box binding factor 2A - s... 400 e-112
TC88990 similar to PIR|T11751|T11751 transcription repressor ROM... 189 1e-48
TC87794 similar to GP|13775107|gb|AAK39130.1 bZIP transcription ... 108 3e-24
TC87793 similar to GP|13775107|gb|AAK39130.1 bZIP transcription ... 101 5e-22
TC91337 weakly similar to GP|13365772|dbj|BAB39174. RISBZ4 {Oryz... 63 2e-10
TC87979 weakly similar to GP|10954097|gb|AAG25728.1 bZIP protein... 58 6e-09
TC81433 similar to GP|13430400|gb|AAK25822.1 bZip transcription ... 57 1e-08
TC85773 weakly similar to GP|9650828|emb|CAC00658.1 common plant... 57 2e-08
TC85772 weakly similar to GP|9650828|emb|CAC00658.1 common plant... 57 2e-08
TC80554 similar to GP|13430400|gb|AAK25822.1 bZip transcription ... 55 7e-08
TC77200 similar to GP|22597162|gb|AAN03468.1 bZIP transcription ... 54 1e-07
TC92502 weakly similar to GP|22597162|gb|AAN03468.1 bZIP transcr... 54 1e-07
TC86844 similar to GP|16797791|gb|AAL27150.1 bZIP transcription ... 53 3e-07
TC81252 similar to PIR|T52624|T52624 b-Zip DNA binding protein [... 50 1e-06
TC78431 similar to GP|16797791|gb|AAL27150.1 bZIP transcription ... 49 4e-06
TC81755 homologue to GP|15100055|gb|AAK84223.1 transcription fac... 49 4e-06
TC87112 similar to GP|2921823|gb|AAC04862.1| shoot-forming PKSF1... 47 1e-05
BF647768 weakly similar to PIR|T03990|T039 probable transcriptio... 47 1e-05
>TC83004 similar to PIR|T10985|T10985 regulator protein ROM2 - kidney bean,
partial (78%)
Length = 1250
Score = 518 bits (1333), Expect(2) = e-155
Identities = 270/379 (71%), Positives = 310/379 (81%), Gaps = 2/379 (0%)
Frame = +1
Query: 48 MPPYYNSPVASGHTPHPYMWGPPQPMMPPYGHPYAA-MYPHGGVYTHPAVPIGPHPHSQG 106
+PPY+NS A GH PHPYMWGPPQPMM PYG PYA Y HGGVYTHPAV IG + + QG
Sbjct: 85 IPPYFNSAAAPGHAPHPYMWGPPQPMMHPYGPPYAPPFYSHGGVYTHPAVAIGSNSNGQG 264
Query: 107 ISSSPATGTPLSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQS 166
ISSSPA GTP SIETP KSSGNTDQGLMKKLKGFDGLAMSIGNG+AESAE GAE+R S+S
Sbjct: 265 ISSSPAAGTPTSIETPTKSSGNTDQGLMKKLKGFDGLAMSIGNGNAESAERGAENRLSRS 444
Query: 167 VNTEGSSDGSDGNTSGANQTRRKRSREGTP-TTDGEGKTNTQGSQISKEIAASDKMMAVA 225
V+TEGSSDGSDGNT+G N T RKRSR+GTP TTDGEGKT SQ+SKE AAS K ++V
Sbjct: 445 VDTEGSSDGSDGNTTGTNGT-RKRSRDGTPTTTDGEGKTEMPDSQVSKETAASKKTVSVI 621
Query: 226 PAGVTGQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRE 285
+ +VGPV SS MTT+LELRN S + TS PQP VLPPEAW+QNERELKRE
Sbjct: 622 TSSAAENMVGPVLSSGMTTSLELRNPSPI-----STSAPQPCGVLPPEAWMQNERELKRE 786
Query: 286 RRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALK 345
RRKQSNRESARRSRLRKQAEAEELAR+V++L AE+ +L+SE+N LAENS +L++ENA LK
Sbjct: 787 RRKQSNRESARRSRLRKQAEAEELARRVDALTAENLALKSEMNELAENSAKLKIENATLK 966
Query: 346 EKFKIAKLGQPKEIILTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDENGYCDNKPNS 405
EK + +LGQ +EIIL +D +R TPVSTENLLSRV N+S S+DR E+ENG+C+NKPNS
Sbjct: 967 EKLENTQLGQTEEIILNGMD-KRATPVSTENLLSRV-NDSNSDDRAAEEENGFCENKPNS 1140
Query: 406 GAKLHQLLDASPRADAVAA 424
GAKL QLLD +PRA+AVAA
Sbjct: 1141GAKLRQLLDTNPRANAVAA 1197
Score = 49.7 bits (117), Expect(2) = e-155
Identities = 20/25 (80%), Positives = 22/25 (88%)
Frame = +2
Query: 22 DQTNQTNVHVYPDWAAMQAYYGPRV 46
DQ N +N+HVYPDWAAMQAYYG RV
Sbjct: 8 DQANPSNMHVYPDWAAMQAYYGQRV 82
>TC78777 similar to PIR|T10985|T10985 regulator protein ROM2 - kidney bean,
partial (84%)
Length = 1822
Score = 437 bits (1123), Expect(2) = e-127
Identities = 227/227 (100%), Positives = 227/227 (100%)
Frame = +1
Query: 199 DGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKT 258
DGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKT
Sbjct: 745 DGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKT 924
Query: 259 NPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNA 318
NPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNA
Sbjct: 925 NPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNA 1104
Query: 319 ESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTTPVSTENLL 378
ESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTTPVSTENLL
Sbjct: 1105ESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTTPVSTENLL 1284
Query: 379 SRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDASPRADAVAAG 425
SRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDASPRADAVAAG
Sbjct: 1285SRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDASPRADAVAAG 1425
Score = 357 bits (917), Expect = 4e-99
Identities = 165/165 (100%), Positives = 165/165 (100%)
Frame = +3
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH 60
MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH
Sbjct: 63 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH 242
Query: 61 TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 120
TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE
Sbjct: 243 TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 422
Query: 121 TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQ 165
TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQ
Sbjct: 423 TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQ 557
Score = 38.5 bits (88), Expect(2) = e-127
Identities = 17/23 (73%), Positives = 20/23 (86%)
Frame = +2
Query: 166 SVNTEGSSDGSDGNTSGANQTRR 188
SVNTEGSSDGSDGNTSG + ++
Sbjct: 695 SVNTEGSSDGSDGNTSGMEKGKQ 763
>TC78776 similar to PIR|T07150|T07150 G-box binding factor 2A - soybean
(fragment), partial (39%)
Length = 691
Score = 400 bits (1029), Expect = e-112
Identities = 187/188 (99%), Positives = 187/188 (99%)
Frame = +2
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH 60
MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH
Sbjct: 128 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH 307
Query: 61 TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 120
TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE
Sbjct: 308 TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 487
Query: 121 TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNT 180
TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNT
Sbjct: 488 TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNT 667
Query: 181 SGANQTRR 188
SGANQ RR
Sbjct: 668 SGANQNRR 691
>TC88990 similar to PIR|T11751|T11751 transcription repressor ROM1 - kidney
bean, partial (91%)
Length = 1528
Score = 189 bits (481), Expect = 1e-48
Identities = 134/354 (37%), Positives = 183/354 (50%), Gaps = 13/354 (3%)
Frame = +1
Query: 6 EEKSTKTEKPSSPVTVDQTNQTNVHVYPDWA-AMQAYYGPRVAMPPYYNSPVASGHTPHP 64
E+ +TK K SS T + T YPDW+ +MQAYY P A PPYY S VAS TPHP
Sbjct: 220 EDSTTKPSKTSSS-TQEVPTPTVQPSYPDWSTSMQAYYNPGAAPPPYYASTVASP-TPHP 393
Query: 65 YMWGPPQPMMPPYGHP--YAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIETP 122
YMWG PMM PYG P Y AM+P G +Y HP++ + P Q + E
Sbjct: 394 YMWGGQHPMMAPYGTPVPYPAMFPPGNIYAHPSMVVTPSAMHQ------------TTEFE 537
Query: 123 PKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSD-GSDG--N 179
K D+ KK KG + N A++ E G S + S D GS+G N
Sbjct: 538 GKGPDGKDKDSSKKPKG------TSANTSAKAGEGGKAGSGSGNDGFSHSGDSGSEGSSN 699
Query: 180 TSGANQTRRKRSREGT---PTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGP 236
S NQ R+++G+ DG N IS+ + V G V
Sbjct: 700 ASDENQQESARNKKGSFDLMLVDGANAQNNTTGPISQ-------------SSVPGNPVVS 840
Query: 237 VASSAMTTALELRNSSSVHSKTNPTSTPQPSAV---LPPEAWIQ-NERELKRERRKQSNR 292
+ ++ + ++L N+SS ++ QP A E W+Q ++RELKR++RKQSNR
Sbjct: 841 IPATNLNIGMDLWNASSAGAEAAKMRHNQPGAPGAGALGEQWMQQDDRELKRQKRKQSNR 1020
Query: 293 ESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKE 346
ESAR+SRLRKQAE EEL ++VE+L E+ +LR E+ +L+E E+L EN ++KE
Sbjct: 1021ESARKSRLRKQAECEELLKRVEALGGENRTLREELQKLSEECEKLTSENDSIKE 1182
>TC87794 similar to GP|13775107|gb|AAK39130.1 bZIP transcription factor 2
{Phaseolus vulgaris}, partial (50%)
Length = 1264
Score = 108 bits (271), Expect = 3e-24
Identities = 78/204 (38%), Positives = 113/204 (55%), Gaps = 17/204 (8%)
Frame = +2
Query: 172 SSDGSDGNTSGANQTR---RKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAG 228
+S+GSD N+ +Q + R+ S E P+ +G Q ++ ++ M V P
Sbjct: 14 ASEGSDENSQNGSQLKFGERQDSFEDDPSQNGSSVP--QNGALNTPHTVVNQTMXVVPMS 187
Query: 229 VTGQLV---GPVASSAMTTALELRNSSS----VHSKTNPTSTPQ------PSAVLPPEAW 275
V G L GP + + +SS +H K T+ P + + W
Sbjct: 188 VAGPLTTVPGPTTNLNIGMDYWGTPTSSTIPAMHGKVPSTAVAGGMVNAGPRDGVHSQLW 367
Query: 276 IQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSE 335
+Q+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ E LN E+ASLR+E++ + E
Sbjct: 368 LQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNQENASLRAELSPIKSEYE 547
Query: 336 RLRMENAALKEKFKIAKLGQ-PKE 358
++ ENA+LKE +LG+ PKE
Sbjct: 548 KIPSENASLKE-----RLGEIPKE 604
>TC87793 similar to GP|13775107|gb|AAK39130.1 bZIP transcription factor 2
{Phaseolus vulgaris}, partial (41%)
Length = 1078
Score = 101 bits (252), Expect = 5e-22
Identities = 69/191 (36%), Positives = 100/191 (52%), Gaps = 15/191 (7%)
Frame = +1
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVH-----VYPDWAAMQAYYGPRVAMPPYYNSP 55
M ++D +K+ K ++P +P Q++ V P+WA QAY P + P +
Sbjct: 280 MSSNDVDKTIKEKEPKTPPAATSQEQSSTTTGTPAVNPEWANYQAY--PSIPPPGF---- 441
Query: 56 VASGHTPHPYMWGPPQPMMPPYG---HPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSP- 111
+AS HPYMWG Q MMPPYG HPY AMYPHGG+Y HP++P G +P S SP
Sbjct: 442 MASSPQAHPYMWGV-QHMMPPYGTPPHPYVAMYPHGGIYAHPSMPPGSYPFSPYAMPSPN 618
Query: 112 ----ATG-TPLSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAE-PGAESRQSQ 165
A+G TP S E K ++ +K+ KG G + + + E + PGA +
Sbjct: 619 GMVDASGNTPGSSEADGKPHEVKEKLPIKRSKGSLGSSNMVTRKNNELGKTPGASANGIH 798
Query: 166 SVNTEGSSDGS 176
S + E +S+G+
Sbjct: 799 SKSGESASEGT 831
>TC91337 weakly similar to GP|13365772|dbj|BAB39174. RISBZ4 {Oryza sativa},
partial (38%)
Length = 1112
Score = 62.8 bits (151), Expect = 2e-10
Identities = 48/166 (28%), Positives = 80/166 (47%), Gaps = 10/166 (6%)
Frame = +1
Query: 212 SKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQPSAV-- 269
S+ + + + + G + PV+++ NS H K + + PS
Sbjct: 217 SQNFTPKHSTITASQSSIYGTVGSPVSANKP-------NSRENHIKGTASGSSDPSDEDN 375
Query: 270 -LPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEIN 328
P I N ++KR+RRK SN ESARRSR RKQA EL +VE L E+A+L +
Sbjct: 376 ESGPCEQITNPVDMKRQRRKDSNCESARRSRWRKQAHLSELEAQVEKLKLENATLYKQFT 555
Query: 329 RLAE-------NSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQ 367
++ N+ L+ + AL+ K K+A+ + + T++++Q
Sbjct: 556 DTSQQFHEADTNNRVLKSDVEALRAKVKLAEDMVTRSSLTTSLNNQ 693
>TC87979 weakly similar to GP|10954097|gb|AAG25728.1 bZIP protein BZO2H2
{Arabidopsis thaliana}, partial (49%)
Length = 1346
Score = 58.2 bits (139), Expect = 6e-09
Identities = 55/172 (31%), Positives = 79/172 (44%), Gaps = 24/172 (13%)
Frame = +2
Query: 229 VTGQLVGPVASSAMTTALELRN--SSSVHSKTNPTSTPQPSAVLP--------PEAWIQN 278
+T + + SA T+ N SSS ++T +T S+ P P N
Sbjct: 374 ITATIESQSSISATVTSPVSANKPSSSRENQTKGVTTTSGSSRDPSDEDDEAGPCEQSTN 553
Query: 279 ERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAE------ 332
++KR RRK SNRESARRSR RKQA +L +VE L E+ASL ++ ++
Sbjct: 554 PVDMKRLRRKVSNRESARRSRRRKQAHLADLEVQVEQLRLENASLFKQLTDASQQFRDAN 733
Query: 333 -NSERLRMENAALKEKFKIAK-------LGQPKEIILTNIDSQRTTPVSTEN 376
N+ L+ + AL+ K K+A+ L +L N TTP N
Sbjct: 734 TNNRVLKSDVEALRAKVKLAEDMVSRGTLPTFNNQLLQNQSQLNTTPPQINN 889
>TC81433 similar to GP|13430400|gb|AAK25822.1 bZip transcription factor
{Phaseolus vulgaris}, partial (68%)
Length = 774
Score = 57.4 bits (137), Expect = 1e-08
Identities = 39/111 (35%), Positives = 62/111 (55%), Gaps = 2/111 (1%)
Frame = +2
Query: 239 SSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEA-WIQ-NERELKRERRKQSNRESAR 296
S+ + T S+ H + P S ++ EA +Q N + ++ RR SNRESAR
Sbjct: 137 SNILGTLPNFHYPSAGHDQFAPPSCLSSNSTTSDEADELQFNIIDERKHRRMISNRESAR 316
Query: 297 RSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEK 347
RSR+RKQ +EL +V L E+ +L ++N ++E+ + + ENA LKE+
Sbjct: 317 RSRMRKQKHLDELWSQVMKLRTENHNLVDKLNHVSESHDTVVQENARLKEE 469
>TC85773 weakly similar to GP|9650828|emb|CAC00658.1 common plant regulatory
factor 7 {Petroselinum crispum}, partial (62%)
Length = 945
Score = 56.6 bits (135), Expect = 2e-08
Identities = 43/148 (29%), Positives = 77/148 (51%), Gaps = 2/148 (1%)
Frame = +1
Query: 244 TALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQ-NERELKRERRKQSNRESARRSRLRK 302
TA+++ +S +S + TS+ Q S +Q N + K+ +R QSNRESARRSR++K
Sbjct: 154 TAIDMASSGGTYS--SGTSSLQNSGSEGDHNHVQVNITDQKKRKRMQSNRESARRSRMKK 327
Query: 303 QAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALK-EKFKIAKLGQPKEIIL 361
Q E+L+ ++E L E+ + + + + + ENA L+ + +++ Q I+
Sbjct: 328 QQHMEDLSNQIEQLKKENIQISTNVGVTTQMYLNVESENAILRVQMAELSHRLQSLNDII 507
Query: 362 TNIDSQRTTPVSTENLLSRVNNNSGSND 389
I+S + T+ L N+ G +D
Sbjct: 508 HYIESSNSLFQETDQLF----NDCGFSD 579
>TC85772 weakly similar to GP|9650828|emb|CAC00658.1 common plant regulatory
factor 7 {Petroselinum crispum}, partial (64%)
Length = 1224
Score = 56.6 bits (135), Expect = 2e-08
Identities = 43/148 (29%), Positives = 77/148 (51%), Gaps = 2/148 (1%)
Frame = +1
Query: 244 TALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQ-NERELKRERRKQSNRESARRSRLRK 302
TA+++ +S +S + TS+ Q S +Q N + K+ +R QSNRESARRSR++K
Sbjct: 643 TAIDMASSGGTYS--SGTSSLQNSGSEGDHNHVQVNITDQKKRKRMQSNRESARRSRMKK 816
Query: 303 QAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALK-EKFKIAKLGQPKEIIL 361
Q E+L+ ++E L E+ + + + + + ENA L+ + +++ Q I+
Sbjct: 817 QQHMEDLSNQIEQLKKENIQISTNVGVTTQMYLNVESENAILRVQMAELSHRLQSLNDII 996
Query: 362 TNIDSQRTTPVSTENLLSRVNNNSGSND 389
I+S + T+ L N+ G +D
Sbjct: 997 HYIESSNSLFQETDQLF----NDCGFSD 1068
>TC80554 similar to GP|13430400|gb|AAK25822.1 bZip transcription factor
{Phaseolus vulgaris}, partial (52%)
Length = 839
Score = 54.7 bits (130), Expect = 7e-08
Identities = 28/65 (43%), Positives = 41/65 (63%)
Frame = +1
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
++ RR SNRESARRSR+RKQ + +EL +V L E+ L ++N E +++ EN
Sbjct: 307 RKHRRMVSNRESARRSRMRKQKQLDELWSQVVWLRNENHQLLDKLNNFCETHDKVVQENV 486
Query: 343 ALKEK 347
LKE+
Sbjct: 487 QLKEQ 501
>TC77200 similar to GP|22597162|gb|AAN03468.1 bZIP transcription factor ATB2
{Glycine max}, partial (84%)
Length = 1481
Score = 53.9 bits (128), Expect = 1e-07
Identities = 37/141 (26%), Positives = 71/141 (50%)
Frame = +1
Query: 276 IQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSE 335
+Q + ++ +R SNRESARRSR+RKQ ++L +V L E+ + + +N +
Sbjct: 598 LQALMDQRKRKRMISNRESARRSRMRKQKHLDDLVSQVSKLRKENQEILTSVNITTQKYL 777
Query: 336 RLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDE 395
+ EN+ L+ A++G+ + ++ + L+ N G+++ VE
Sbjct: 778 SVEAENSVLR-----AQMGE--------LSNRLESLNEIVGALNSSNGVFGASNAFVEQN 918
Query: 396 NGYCDNKPNSGAKLHQLLDAS 416
NG+ N N+ + ++Q + AS
Sbjct: 919 NGFFFNSLNNMSYMNQPIMAS 981
>TC92502 weakly similar to GP|22597162|gb|AAN03468.1 bZIP transcription
factor ATB2 {Glycine max}, partial (59%)
Length = 597
Score = 53.9 bits (128), Expect = 1e-07
Identities = 25/70 (35%), Positives = 44/70 (62%)
Frame = +3
Query: 276 IQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSE 335
+Q + ++ +RKQS RESARRSR+RKQ ++L +VE L E++ + + +N ++
Sbjct: 51 LQRLMDQRKRKRKQSGRESARRSRMRKQKHMDDLIAEVERLRNENSEILTRMNMTTQHYL 230
Query: 336 RLRMENAALK 345
++ EN L+
Sbjct: 231 KIEAENCVLR 260
>TC86844 similar to GP|16797791|gb|AAL27150.1 bZIP transcription factor
{Nicotiana tabacum}, partial (47%)
Length = 1609
Score = 52.8 bits (125), Expect = 3e-07
Identities = 29/68 (42%), Positives = 38/68 (55%)
Frame = +1
Query: 278 NERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERL 337
N + KR RR SNRESARRSR RKQA EL +V L E++SL + + +
Sbjct: 658 NPTDAKRVRRMLSNRESARRSRRRKQAHLTELETQVSELRGENSSLLKRLTDVTQKFNNS 837
Query: 338 RMENAALK 345
++N LK
Sbjct: 838 AVDNRILK 861
>TC81252 similar to PIR|T52624|T52624 b-Zip DNA binding protein [imported] -
Arabidopsis thaliana, partial (48%)
Length = 1221
Score = 50.4 bits (119), Expect = 1e-06
Identities = 41/132 (31%), Positives = 64/132 (48%), Gaps = 9/132 (6%)
Frame = +2
Query: 223 AVAPAGVTGQLVGPVASSAMTTA-LELRNSSSVHSKTNPTST----PQPSAVLPP----E 273
A A +G +G G + + TT R+S SV T+ T+ +PP E
Sbjct: 392 AGAGSGTSGNNDGEKSGNDATTPRTRHRHSCSVDGSTSTTNMFGEIMDAKKAMPPDKLAE 571
Query: 274 AWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAEN 333
W + KR +R +NR+SA RS+ RK +EL RKV++L E+ +L +++ +
Sbjct: 572 LWTIDP---KRAKRILANRQSAARSKERKARYIQELERKVQTLQTEATTLSAQLTLYQRD 742
Query: 334 SERLRMENAALK 345
+ L EN LK
Sbjct: 743 TNGLSTENTELK 778
>TC78431 similar to GP|16797791|gb|AAL27150.1 bZIP transcription factor
{Nicotiana tabacum}, partial (61%)
Length = 1546
Score = 48.9 bits (115), Expect = 4e-06
Identities = 27/65 (41%), Positives = 38/65 (57%)
Frame = +1
Query: 281 ELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRME 340
++KR RR SNRESARRSR RKQA EL +V L E+++L + + + ++
Sbjct: 745 DVKRVRRMLSNRESARRSRRRKQAHLTELETQVSQLRGENSTLVKRLTDVNQKYTDSAVD 924
Query: 341 NAALK 345
N LK
Sbjct: 925 NRVLK 939
>TC81755 homologue to GP|15100055|gb|AAK84223.1 transcription factor bZIP61
{Arabidopsis thaliana}, partial (30%)
Length = 724
Score = 48.9 bits (115), Expect = 4e-06
Identities = 31/75 (41%), Positives = 44/75 (58%)
Frame = +2
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
KR +R +NR+SA+RSR+RK EL R V SL AE + L + L L ++N+
Sbjct: 71 KRVKRILANRQSAQRSRVRKLQYISELERSVTSLQAEVSVLSPRVAYLDHQRLLLNVDNS 250
Query: 343 ALKEKFKIAKLGQPK 357
A+K+ +IA L Q K
Sbjct: 251 AIKQ--RIAALAQDK 289
>TC87112 similar to GP|2921823|gb|AAC04862.1| shoot-forming PKSF1 {Paulownia
kawakamii}, partial (30%)
Length = 1379
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/65 (38%), Positives = 39/65 (59%)
Frame = +1
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
KR +R +NR+SA RS+ RK EL RKV++L E+ +L +++ L ++ L +N
Sbjct: 571 KRAKRILANRQSAARSKERKTRYTSELERKVQTLQTEATNLSAQLTTLQRDTTDLTAQNK 750
Query: 343 ALKEK 347
LK K
Sbjct: 751 ELKMK 765
>BF647768 weakly similar to PIR|T03990|T039 probable transcription activator
- rice, partial (29%)
Length = 660
Score = 47.0 bits (110), Expect = 1e-05
Identities = 31/69 (44%), Positives = 37/69 (52%), Gaps = 3/69 (4%)
Frame = +1
Query: 250 NSSSVHSKTNPTSTPQPSAV---LPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEA 306
NS H K + + PS P I N ++KR+RRK SN ESARRSR RKQA
Sbjct: 13 NSRENHIKGTASGSSDPSDEDNESGPCEQITNPVDMKRQRRKDSNCESARRSRWRKQAHL 192
Query: 307 EELARKVES 315
EL +V S
Sbjct: 193 SELEAQVTS 219
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.305 0.123 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,181,061
Number of Sequences: 36976
Number of extensions: 187532
Number of successful extensions: 2540
Number of sequences better than 10.0: 239
Number of HSP's better than 10.0 without gapping: 1381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1791
length of query: 425
length of database: 9,014,727
effective HSP length: 99
effective length of query: 326
effective length of database: 5,354,103
effective search space: 1745437578
effective search space used: 1745437578
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137602.6


