

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.5 - phase: 0
(865 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
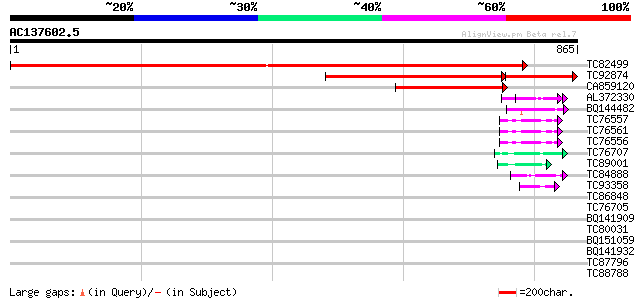
Sequences producing significant alignments: (bits) Value
TC82499 SP|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation f... 1470 0.0
TC92874 PIR|T50775|T50775 probable translation initiation factor... 541 0.0
CA859120 weakly similar to EGAD|56711|A-23 eIF-3 p110 subunit ge... 153 3e-37
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 51 2e-06
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 47 2e-05
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 47 4e-05
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 47 4e-05
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 47 4e-05
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 45 2e-04
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 43 6e-04
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 43 6e-04
TC93358 weakly similar to GP|7303636|gb|AAF58688.1| CG13214-PA {... 42 8e-04
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 42 0.001
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 42 0.001
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 41 0.002
TC80031 similar to GP|21327989|dbj|BAC00578. contains ESTs AU093... 41 0.002
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 40 0.003
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 40 0.004
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 40 0.005
TC88788 weakly similar to SP|P05790|FBOH_BOMMO Fibroin heavy cha... 40 0.005
>TC82499 SP|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation factor 3
subunit 8 (eIF3 p110) (eIF3c). [Barrel medic], partial
(91%)
Length = 2816
Score = 1470 bits (3806), Expect = 0.0
Identities = 746/789 (94%), Positives = 768/789 (96%)
Frame = +1
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
MASTVDQIKNAIKINDWVSLQE+FDKINKQL+KVMRVIESQKIPNLYIKALVMLEDFLAQ
Sbjct: 454 MASTVDQIKNAIKINDWVSLQESFDKINKQLEKVMRVIESQKIPNLYIKALVMLEDFLAQ 633
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
AS+NKDAKKKMSP+NAKAFNSMKQKLKKNNKQYEDLIIK RE+P+SE EK EDDEDSD+Y
Sbjct: 634 ASANKDAKKKMSPSNAKAFNSMKQKLKKNNKQYEDLIIKCRESPESEGEKDEDDEDSDEY 813
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
SDD E+IEPDQL+KPEPVSDSETSELGNDRPGDD DAPWDQKL KKDRLLEKMFMKKPS
Sbjct: 814 ESDD-EMIEPDQLRKPEPVSDSETSELGNDRPGDDGDAPWDQKLSKKDRLLEKMFMKKPS 990
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVN 240
EITWDTVN KFKEILEARGRKGTGRFEQVEQ TFLTKVAKTPAQKLQILFSVVSAQFDVN
Sbjct: 991 EITWDTVNKKFKEILEARGRKGTGRFEQVEQLTFLTKVAKTPAQKLQILFSVVSAQFDVN 1170
Query: 241 PGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYDGPIHVWGN 300
PGLSGHMPI+VWKKCVQNMLVILDILVQHPNIKVDDSVELDENE+KKGDDY+GPI+VWGN
Sbjct: 1171 PGLSGHMPISVWKKCVQNMLVILDILVQHPNIKVDDSVELDENETKKGDDYNGPINVWGN 1350
Query: 301 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGDFKASSKVA 360
LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEP FVVLAQNVQEYLESIGDFKASSKVA
Sbjct: 1351 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPQFVVLAQNVQEYLESIGDFKASSKVA 1530
Query: 361 LKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVTPELVARK 420
LKRVELIYYKPHEVYEATRKLAE+TV+GDNGE +SEE KGF DTRI APFVVT ELVARK
Sbjct: 1531 LKRVELIYYKPHEVYEATRKLAEMTVEGDNGE-MSEEPKGFEDTRIPAPFVVTLELVARK 1707
Query: 421 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 480
PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV
Sbjct: 1708 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 1887
Query: 481 HHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEK 540
HHMDISTQILFNRAMSQLGLCAFR GLVSEAHGCLSELYSGGRVKELLAQGVSQ+RYHEK
Sbjct: 1888 HHMDISTQILFNRAMSQLGLCAFRAGLVSEAHGCLSELYSGGRVKELLAQGVSQSRYHEK 2067
Query: 541 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 600
TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL
Sbjct: 2068 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 2247
Query: 601 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDK 660
EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASL+VWKFVKNRD VLEMLKDK
Sbjct: 2248 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLDVWKFVKNRDAVLEMLKDK 2427
Query: 661 IKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCI 720
IKEEALRTYLFTFSSSYDSLSV QLTNFFDLSLPR HSIVSRMM+NEELHASWDQPTGCI
Sbjct: 2428 IKEEALRTYLFTFSSSYDSLSVVQLTNFFDLSLPRVHSIVSRMMVNEELHASWDQPTGCI 2607
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGG 780
IFRNVE S+VQALAFQLTEKLSILAESNERA+EARLGGGGLDLPP+RRDGQDYAAAAAGG
Sbjct: 2608 IFRNVEHSKVQALAFQLTEKLSILAESNERATEARLGGGGLDLPPKRRDGQDYAAAAAGG 2787
Query: 781 GGGTSSGGR 789
G GTSSGGR
Sbjct: 2788 GSGTSSGGR 2814
>TC92874 PIR|T50775|T50775 probable translation initiation factor [imported]
- barrel medic, complete
Length = 1396
Score = 541 bits (1394), Expect(2) = 0.0
Identities = 276/276 (100%), Positives = 276/276 (100%)
Frame = +2
Query: 482 HMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEKT 541
HMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEKT
Sbjct: 2 HMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEKT 181
Query: 542 PEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLLE 601
PEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLLE
Sbjct: 182 PEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLLE 361
Query: 602 VSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDKI 661
VSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDKI
Sbjct: 362 VSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDKI 541
Query: 662 KEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCII 721
KEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCII
Sbjct: 542 KEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCII 721
Query: 722 FRNVELSRVQALAFQLTEKLSILAESNERASEARLG 757
FRNVELSRVQALAFQLTEKLSILAESNERASEARLG
Sbjct: 722 FRNVELSRVQALAFQLTEKLSILAESNERASEARLG 829
Score = 227 bits (578), Expect(2) = 0.0
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ
Sbjct: 828 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 1007
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRGVRA 865
AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRGVRA
Sbjct: 1008AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRGVRA 1154
>CA859120 weakly similar to EGAD|56711|A-23 eIF-3 p110 subunit gen {Homo
sapiens}, partial (10%)
Length = 659
Score = 153 bits (386), Expect = 3e-37
Identities = 75/171 (43%), Positives = 112/171 (64%)
Frame = +1
Query: 589 RKIISKNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVK 648
++IISK FRR+L+ +E+Q F+GPPE RDH+M A + L G++Q+ ++I+++++W +
Sbjct: 1 QEIISKPFRRMLDFNERQVFSGPPENTRDHIMGAAKALAAGEWQRCQELISAIKIWDLMP 180
Query: 649 NRDTVLEMLKDKIKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEE 708
N + +ML KI+EE LRTYLFT+SS Y +L +DQL N F L + SI+S+M+ NEE
Sbjct: 181 NTLKIKDMLNRKIQEEGLRTYLFTYSSHYVTLGLDQLANMFSLPVNSVTSIISKMIWNEE 360
Query: 709 LHASWDQPTGCIIFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGG 759
L AS DQ ++ VE ++VQ LA EK + E+NER E + G
Sbjct: 361 LAASLDQVDNNVVLHRVEPTKVQQLALSFAEKAASFVEANERTLEQKQTSG 513
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 51.2 bits (121), Expect = 2e-06
Identities = 34/93 (36%), Positives = 39/93 (41%)
Frame = +2
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
+S GG P G Y ++GG GG+S G Y + G G GYGG
Sbjct: 38 SSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGG-------YGGSSGGYGGGGYGGG 196
Query: 811 ALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
A GG GG S G G GGGGY GG
Sbjct: 197 ASGGYGGGGYGGGS-GGGYGGGGYGGGGYGGGG 292
Score = 49.7 bits (117), Expect = 5e-06
Identities = 35/82 (42%), Positives = 42/82 (50%), Gaps = 2/82 (2%)
Frame = +2
Query: 772 DYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY-SRGRGMG 830
DY ++++GG GG+SSGG S S SG GYGG + S G SGGY G G
Sbjct: 2 DYRSSSSGGYGGSSSGGYGGSSSSSYPASSSG-GGYGGGS-SGGYGGSSGGYGGSSGGYG 175
Query: 831 GGGY-QNSSRTQGGSALRGPHG 851
GGGY +S GG G G
Sbjct: 176 GGGYGGGASGGYGGGGYGGGSG 241
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 47.4 bits (111), Expect = 2e-05
Identities = 36/101 (35%), Positives = 42/101 (40%), Gaps = 7/101 (6%)
Frame = -1
Query: 759 GGLDLPPRRRD-GQDYAAAAAG------GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
GG LPP R G+ YAAA GG G SGG + G G GG
Sbjct: 486 GGRSLPPPRASRGRRYAAARCAPVTGPPGGAGQHSGGTGPGGGPGRGGGARGAGGRGGEG 307
Query: 812 LSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGD 852
+A G GG RGRG G + R +GG RG G+
Sbjct: 306 RGQGEAEGGGGGGRGRGAKEG--EGRERGRGGGDRRGGGGE 190
Score = 39.3 bits (90), Expect = 0.007
Identities = 36/102 (35%), Positives = 40/102 (38%), Gaps = 4/102 (3%)
Frame = -3
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGG-GGTSSGGRWQDLSYSQTRQGSGRAGYGG 809
A A G P RDG A A GGG GG GGR R+G G G GG
Sbjct: 430 AMRASDGAARRGRPTLGRDGAGRGAGAGGGGAGGRGPGGR-----RPGPRRG*GGGG-GG 269
Query: 810 RALSFNQAGGSG---GYSRGRGMGGGGYQNSSRTQGGSALRG 848
+ GG G G R +G GGG + GG RG
Sbjct: 268 EGEGGERGGGEGEGEGGRR*KGRGGGKGGRGATLGGGGRSRG 143
Score = 37.7 bits (86), Expect = 0.019
Identities = 34/103 (33%), Positives = 38/103 (36%), Gaps = 1/103 (0%)
Frame = -2
Query: 750 RASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG 809
R +AR G P R G+ A GGGGG G G +AG
Sbjct: 437 RRRDARQ*RGRPAGPANTRAGRG-RAGGRGGGGGRGGPG-----------AGGAKAGAKE 294
Query: 810 RALSFNQAGGSGGYSRGR-GMGGGGYQNSSRTQGGSALRGPHG 851
R GG GG RGR G GGGG + G RG G
Sbjct: 293 RLRGGGGGGGGGGRKRGRGGRGGGGAEIEGEGGGKRWARGDFG 165
Score = 33.1 bits (74), Expect = 0.47
Identities = 28/93 (30%), Positives = 35/93 (37%)
Frame = -3
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G+D A AG GGG G+G G GGR + G GG G G
Sbjct: 382 GRDGAGRGAGAGGG-----------------GAGGRGPGGRRPGPRRG*GGGGGGEGEGG 254
Query: 830 GGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRG 862
GG + +GG +G G R +L G
Sbjct: 253 ERGGGEGEG--EGGRR*KGRGGGKGGRGATLGG 161
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 46.6 bits (109), Expect = 4e-05
Identities = 36/96 (37%), Positives = 40/96 (41%)
Frame = +1
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
N+ S GGGG R G Y GGGG GG + G G GY
Sbjct: 481 NQAQSRGSGGGGG-----GGRGGGGY------GGGGGGYGGERRGYGGGGGYGGGGGGGY 627
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G R GG GGYSRG GGGGY ++GG
Sbjct: 628 GERR------GGGGGYSRGG--GGGGYGGGGYSRGG 711
Score = 35.8 bits (81), Expect = 0.073
Identities = 29/78 (37%), Positives = 33/78 (42%), Gaps = 9/78 (11%)
Frame = +1
Query: 783 GTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY---------SRGRGMGGGG 833
G GR ++ +Q+R GSG G GGR GG GGY G G GGGG
Sbjct: 448 GQDLDGRNITVNQAQSR-GSGGGGGGGRG-GGGYGGGGGGYGGERRGYGGGGGYGGGGGG 621
Query: 834 YQNSSRTQGGSALRGPHG 851
R GG RG G
Sbjct: 622 GYGERRGGGGGYSRGGGG 675
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 46.6 bits (109), Expect = 4e-05
Identities = 36/96 (37%), Positives = 40/96 (41%)
Frame = +1
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
N+ S GGGG R G Y GGGG GG + G G GY
Sbjct: 577 NQAQSRGSGGGGG-----GGRGGGGY------GGGGGGYGGERRGYGGGGGYGGGGGGGY 723
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G R GG GGYSRG GGGGY ++GG
Sbjct: 724 GERR------GGGGGYSRGG--GGGGYGGGGYSRGG 807
Score = 35.8 bits (81), Expect = 0.073
Identities = 29/78 (37%), Positives = 33/78 (42%), Gaps = 9/78 (11%)
Frame = +1
Query: 783 GTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY---------SRGRGMGGGG 833
G GR ++ +Q+R GSG G GGR GG GGY G G GGGG
Sbjct: 544 GQDLDGRNITVNQAQSR-GSGGGGGGGRG-GGGYGGGGGGYGGERRGYGGGGGYGGGGGG 717
Query: 834 YQNSSRTQGGSALRGPHG 851
R GG RG G
Sbjct: 718 GYGERRGGGGGYSRGGGG 771
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 46.6 bits (109), Expect = 4e-05
Identities = 36/96 (37%), Positives = 40/96 (41%)
Frame = +2
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
N+ S GGGG R G Y GGGG GG + G G GY
Sbjct: 323 NQAQSRGSGGGGG-----GGRGGGGY------GGGGGGYGGERRGYGGGGGYGGGGGGGY 469
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G R GG GGYSRG GGGGY ++GG
Sbjct: 470 GERR------GGGGGYSRGG--GGGGYGGGGYSRGG 553
Score = 35.8 bits (81), Expect = 0.073
Identities = 29/78 (37%), Positives = 33/78 (42%), Gaps = 9/78 (11%)
Frame = +2
Query: 783 GTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY---------SRGRGMGGGG 833
G GR ++ +Q+R GSG G GGR GG GGY G G GGGG
Sbjct: 290 GQDLDGRNITVNQAQSR-GSGGGGGGGRG-GGGYGGGGGGYGGERRGYGGGGGYGGGGGG 463
Query: 834 YQNSSRTQGGSALRGPHG 851
R GG RG G
Sbjct: 464 GYGERRGGGGGYSRGGGG 517
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 44.7 bits (104), Expect = 2e-04
Identities = 34/118 (28%), Positives = 48/118 (39%), Gaps = 6/118 (5%)
Frame = +1
Query: 740 KLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTR 799
K ++ ++NE ++A+ GG G G ++ GGG GG + +
Sbjct: 109 KKEVVEKTNE-VNDAKYGGYG--------GGYNHGGGGYNGGGYNHGGGGYNNGGGGYNH 261
Query: 800 QGSGRAGYGGRALSFNQAGGS---GGYSRGRG---MGGGGYQNSSRTQGGSALRGPHG 851
G G GG +N GG GGY+ G G GGGGY + G G HG
Sbjct: 262 GGGGYNNGGG---GYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHG 426
Score = 39.7 bits (91), Expect = 0.005
Identities = 32/97 (32%), Positives = 38/97 (38%), Gaps = 10/97 (10%)
Frame = +1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAG---GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS 813
GGGG + G Y G GGGG ++GG Y+ G GY
Sbjct: 184 GGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGG----GYNHGGGGYNGGGYNHGGGG 351
Query: 814 FNQAGGS-----GGYSRGRGMGG--GGYQNSSRTQGG 843
+N GG GGY+ G G GG GG N GG
Sbjct: 352 YNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGG 462
Score = 37.7 bits (86), Expect = 0.019
Identities = 34/118 (28%), Positives = 40/118 (33%), Gaps = 23/118 (19%)
Frame = +1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAG---GGGGTSSGGRWQDLSYSQTRQGSGRAGYGG---- 809
GGGG + G Y G GGGG + GG G G G+G
Sbjct: 304 GGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESV 483
Query: 810 ------RALSFNQAGGSGGYSRGRGM----------GGGGYQNSSRTQGGSALRGPHG 851
+ N A GGY+ G G GGGGY + GG G HG
Sbjct: 484 AVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHG-GGGHG 654
Score = 29.6 bits (65), Expect = 5.2
Identities = 19/61 (31%), Positives = 24/61 (39%)
Frame = +1
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRT 840
GGG + GG + G G +GG GG GG+ G G G Q +T
Sbjct: 532 GGGYNHGGGSYNHGGGSYHHGGGGYNHGG--------GGHGGHGGGGHGGHGAEQTEDKT 687
Query: 841 Q 841
Q
Sbjct: 688 Q 690
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 42.7 bits (99), Expect = 6e-04
Identities = 29/82 (35%), Positives = 33/82 (39%)
Frame = +3
Query: 745 AESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGR 804
A+S R GGGG G GGGGG GG +D YS++ G G
Sbjct: 279 AQSRGSGGGGRGGGGG---------GYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGY 431
Query: 805 AGYGGRALSFNQAGGSGGYSRG 826
G G R GG GGY G
Sbjct: 432 GGGGDRGYG---GGGGGGYGGG 488
Score = 37.7 bits (86), Expect = 0.019
Identities = 31/94 (32%), Positives = 37/94 (38%), Gaps = 2/94 (2%)
Frame = +3
Query: 760 GLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGG 819
G D+ R + + +GGGG GG + G GYGG + GG
Sbjct: 240 GQDIDGRNITVNEAQSRGSGGGGRGGGGGGY-----------GGGGGYGGGGGGYGGGGG 386
Query: 820 --SGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GGYSR GGGGY GG RG G
Sbjct: 387 RRDGGYSRSG--GGGGY-------GGGGDRGYGG 461
Score = 37.0 bits (84), Expect = 0.033
Identities = 25/70 (35%), Positives = 30/70 (42%)
Frame = +3
Query: 783 GTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQG 842
G GR ++ +Q+R GSG G GG + GG GG G G GGG G
Sbjct: 240 GQDIDGRNITVNEAQSR-GSGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSG 416
Query: 843 GSALRGPHGD 852
G G GD
Sbjct: 417 GGGGYGGGGD 446
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 42.7 bits (99), Expect = 6e-04
Identities = 32/89 (35%), Positives = 38/89 (41%), Gaps = 1/89 (1%)
Frame = +2
Query: 764 PPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAG-GSGG 822
PP+ Q+ GG GG GG + D Y G G YGG G G GG
Sbjct: 269 PPQGAPRQEMGYGGGGGRGGGGYGGGY-DQGY-----GGGGGSYGGGFGGGRGGGRGGGG 430
Query: 823 YSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
Y +G GGGGY QGG +G +G
Sbjct: 431 YXQGGYGGGGGY-----XQGGYGGQGGYG 502
Score = 41.6 bits (96), Expect = 0.001
Identities = 30/88 (34%), Positives = 36/88 (40%), Gaps = 5/88 (5%)
Frame = +2
Query: 769 DGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA-LSFNQAGGSGGY---- 823
DG+ A G G G Q++ Y G G GYGG + GGS G
Sbjct: 224 DGRTKAIDVTGPNGAPPQGAPRQEMGYGGGG-GRGGGGYGGGYDQGYGGGGGSYGGGFGG 400
Query: 824 SRGRGMGGGGYQNSSRTQGGSALRGPHG 851
RG G GGGGY GG +G +G
Sbjct: 401 GRGGGRGGGGYXQGGYGGGGGYXQGGYG 484
>TC93358 weakly similar to GP|7303636|gb|AAF58688.1| CG13214-PA {Drosophila
melanogaster}, partial (7%)
Length = 611
Score = 42.4 bits (98), Expect = 8e-04
Identities = 25/60 (41%), Positives = 31/60 (51%)
Frame = +3
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
G GGG G R+ + S + SGR G GG GG+ G+S GRG GGGG + S
Sbjct: 447 GRGGGGGFGPRFGNGHASDRKYDSGRGGRGG------GRGGAQGFSGGRGRGGGGGRGGS 608
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 42.0 bits (97), Expect = 0.001
Identities = 30/83 (36%), Positives = 35/83 (42%), Gaps = 6/83 (7%)
Frame = +3
Query: 774 AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG----RALSFNQAGG--SGGYSRGR 827
A G GGG GGR D + +G GR G+GG R F GG GG GR
Sbjct: 60 APPVRGRGGGGFRGGRGGD----RGGRGGGRGGFGGRGGDRGTPFKARGGGRGGGGRGGR 227
Query: 828 GMGGGGYQNSSRTQGGSALRGPH 850
G G GG + G + PH
Sbjct: 228 GGGRGGGRGGGMKGGSKVIVEPH 296
Score = 33.1 bits (74), Expect = 0.47
Identities = 25/74 (33%), Positives = 28/74 (37%), Gaps = 5/74 (6%)
Frame = +3
Query: 764 PPRRRDGQDYAAAAAG-----GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAG 818
P R R G + G GGG GGR D +G GR G G + G
Sbjct: 66 PVRGRGGGGFRGGRGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRGGGGRGGRGGGRGG 245
Query: 819 GSGGYSRGRGMGGG 832
G GG GM GG
Sbjct: 246 GRGG-----GMKGG 272
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 41.6 bits (96), Expect = 0.001
Identities = 33/105 (31%), Positives = 45/105 (42%), Gaps = 3/105 (2%)
Frame = +2
Query: 740 KLSILAESNERASEARLGG---GGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYS 796
K ++ ++NE ++A+ GG GG + G Y GGG +GG + +
Sbjct: 149 KKEVVEKTNE-VNDAKYGGYNHGGYN------HGGGYNGGGYNHGGGYHNGGGYHNGGGG 307
Query: 797 QTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQ 841
G G G GG GG GGY+RG G GG G S Q
Sbjct: 308 YHNGGGGYNGGGGH-------GGHGGYNRGGGHGGRGAAESVAVQ 421
Score = 33.9 bits (76), Expect = 0.28
Identities = 35/106 (33%), Positives = 42/106 (39%), Gaps = 7/106 (6%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYS-QTRQGSGR---AGYGGRAL 812
GGGG +G G G GGR S + QT + + A GG
Sbjct: 296 GGGGYHNGGGGYNGGGGHGGHGGYNRGGGHGGRGAAESVAVQTEEKTNEVNDARNGGGGG 475
Query: 813 SFNQAGGSGGYSRGRGM---GGGGYQNSSRTQGGSALRGPHGDVST 855
SFN+ G S Y+ GRG G GGY + GG G HG T
Sbjct: 476 SFNKGGAS--YNHGRGSYHHG*GGYNHGGGGHGGGG-HGSHGAEQT 604
Score = 32.7 bits (73), Expect = 0.61
Identities = 22/58 (37%), Positives = 24/58 (40%), Gaps = 11/58 (18%)
Frame = +2
Query: 805 AGYGG-RALSFNQAGG--SGGYSRGRGM--------GGGGYQNSSRTQGGSALRGPHG 851
A YGG +N GG GGY+ G G GGGGY N G G HG
Sbjct: 188 AKYGGYNHGGYNHGGGYNGGGYNHGGGYHNGGGYHNGGGGYHNGGGGYNGGGGHGGHG 361
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 41.2 bits (95), Expect = 0.002
Identities = 37/124 (29%), Positives = 45/124 (35%), Gaps = 2/124 (1%)
Frame = +2
Query: 735 FQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLS 794
F T S N + + G G RR G+ G GGG + GG
Sbjct: 65 FLFTISFS*WENKNNGRNTTKEGRRGRGSGKMRRRGERGKRGEGGEGGGGAEGG------ 226
Query: 795 YSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM--GGGGYQNSSRTQGGSALRGPHGD 852
+ +G GR G GGR GG RGRG GGG R +GG G G
Sbjct: 227 ---SERGRGRRGGGGR----------GGGRRGRGKKGGGGTKGRGKRERGGGTEGGGKGK 367
Query: 853 VSTR 856
+ R
Sbjct: 368 IEGR 379
>TC80031 similar to GP|21327989|dbj|BAC00578. contains ESTs AU093876(E1018)
AU093877(E1018)~unknown protein, partial (8%)
Length = 902
Score = 40.8 bits (94), Expect = 0.002
Identities = 40/109 (36%), Positives = 43/109 (38%), Gaps = 23/109 (21%)
Frame = +2
Query: 766 RRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG--GRALSFNQAGGSGGY 823
R R G G GGG GGR +Y T GR GYG GR GG GGY
Sbjct: 203 RGRGGAMAPGGFRGRGGGPGHGGRGGYNNYGPT---PGRGGYGPQGRGGYGTPPGGRGGY 373
Query: 824 S---RG----RGMGGG-----GYQN---------SSRTQGGSALRGPHG 851
RG GM GG GYQN S G+ L GP+G
Sbjct: 374 GPPPRGGYGPNGMRGGRPPPPGYQNGPAPYDRRPSPANNYGAPLAGPYG 520
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 40.4 bits (93), Expect = 0.003
Identities = 29/88 (32%), Positives = 30/88 (33%)
Frame = +1
Query: 764 PPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY 823
PP R + GGGGG GG G G G GG GG GG
Sbjct: 13 PPFRGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 192
Query: 824 SRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G G GGGG GG G G
Sbjct: 193 GGGGGGGGGGGGGGGGGGGGGGGGGGGG 276
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 40.0 bits (92), Expect = 0.004
Identities = 39/109 (35%), Positives = 44/109 (39%), Gaps = 12/109 (11%)
Frame = +2
Query: 749 ERASEARLGGGGLDLPP---RRRDGQDYAAAAAGGGGGTSSGG--RWQDLSYSQ--TRQG 801
E E LGGGG R G + GGGGGT+ GG +W + + R G
Sbjct: 194 EGEGEGWLGGGGKRKGRGGGETRAGGEDGGGVGGGGGGTAGGGIPQWVQRAEGEWNCRCG 373
Query: 802 SGR----AGYGGRALSFNQAGGSGGYSRG-RGMGGGGYQNSSRTQGGSA 845
G G GR GG GG S G RG GGG R GG A
Sbjct: 374 GGARVDDRGRPGRGGEVGGWGGGGGESEGARGWGGG------RDGGGQA 502
Score = 35.8 bits (81), Expect = 0.073
Identities = 34/104 (32%), Positives = 41/104 (38%), Gaps = 13/104 (12%)
Frame = +2
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY----GGRALS 813
GGG + R R G+ GGGGG S G R + R G G+AG R
Sbjct: 371 GGGARVDDRGRPGRGGEVGGWGGGGGESEGAR----GWGGGRDGGGQAGVWLVGRFRVAG 538
Query: 814 FNQAGGSGGYSRGRGMGGG---------GYQNSSRTQGGSALRG 848
AGG G + M GG GY +SRT+ G G
Sbjct: 539 DWDAGGLGWW----WMAGGALPVPKNEVGYCMTSRTRPGRGRAG 658
Score = 33.9 bits (76), Expect = 0.28
Identities = 35/97 (36%), Positives = 41/97 (42%), Gaps = 11/97 (11%)
Frame = +3
Query: 750 RASEARLGGGGLDLPPRRRDGQDYAAAAAG-------GGGGTSSGGRWQDLSYSQTRQGS 802
R E+R G G RR G AA G GGGG S GG + ++ R G
Sbjct: 312 RGGESRNGYKG-----RRGSGTADAAGGRGSMTVGDRGGGGRSEGGGAEG-GRAKARVGG 473
Query: 803 GRAGYGGRALSFNQAGGSGGYSRGRGM----GGGGYQ 835
G G GG GSG RG GM GGGG++
Sbjct: 474 GADGMGGGRQGCGWWVGSGW--RGIGMLGG*GGGGWR 578
Score = 32.7 bits (73), Expect = 0.61
Identities = 25/70 (35%), Positives = 30/70 (42%), Gaps = 4/70 (5%)
Frame = +3
Query: 778 AGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG----GRALSFNQAGGSGGYSRGRGMGGGG 833
AGGGGG GGR G GRAG G GR + + GG G GGGG
Sbjct: 162 AGGGGGGVGGGR-----------GRGRAG*GEEERGRGVG-GERRELGGRMEGEWGGGGG 305
Query: 834 YQNSSRTQGG 843
+ ++ G
Sbjct: 306 GRRGGESRNG 335
Score = 32.3 bits (72), Expect = 0.80
Identities = 25/82 (30%), Positives = 33/82 (39%)
Frame = +1
Query: 768 RDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGR 827
R G+ GGGGG G R ++ +GGR S+ GG G G
Sbjct: 163 RGGEGGGWGGGGGGGGLVRGRRKEE------------GAWGGRDESWG--GGWRGSGGGG 300
Query: 828 GMGGGGYQNSSRTQGGSALRGP 849
G GGG + T+GG + P
Sbjct: 301 GGDGGGGNPAMGTKGGGGVELP 366
Score = 28.9 bits (63), Expect = 8.9
Identities = 18/49 (36%), Positives = 20/49 (40%)
Frame = +2
Query: 803 GRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G G GGR GG GG G G GGG + R G + G G
Sbjct: 152 GCGGGGGRG------GGGGGEGEGEGWLGGGGKRKGRGGGETRAGGEDG 280
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 39.7 bits (91), Expect = 0.005
Identities = 29/83 (34%), Positives = 35/83 (41%), Gaps = 1/83 (1%)
Frame = +1
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G+D+ GGGG GG ++ GSG G GG + GG GG G G
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGGAHG-VGYGSG-GGTGGGYGGGSPGGGGGGGGSGGGG 174
Query: 830 GGGGYQNSSRTQG-GSALRGPHG 851
GGGG + G G G HG
Sbjct: 175 GGGGAHGGAYGGGIGGGEGGGHG 243
>TC88788 weakly similar to SP|P05790|FBOH_BOMMO Fibroin heavy chain
precursor (Fib-H) (H-fibroin). [Silk moth] {Bombyx
mori}, partial (2%)
Length = 782
Score = 39.7 bits (91), Expect = 0.005
Identities = 36/124 (29%), Positives = 46/124 (37%), Gaps = 9/124 (7%)
Frame = +3
Query: 730 VQALAFQLTEKLSILAESNERASEAR---------LGGGGLDLPPRRRDGQDYAAAAAGG 780
V + ++ +S+L N A E+R LGG G+ L G AG
Sbjct: 54 VTMASMKILAVVSLLVAMNVIACESRAARKDLGLDLGGVGIGLGAGLGLGLG-GGGGAGS 230
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRT 840
G G SG S S + GSG +G G A S + G G G G Y S
Sbjct: 231 GAGAGSGSGSGSASTSGSGSGSGSSGAGSEAGSHAGSRAGSGAGSGAGSEAGSYAGSHAG 410
Query: 841 QGGS 844
G S
Sbjct: 411 SGSS 422
Score = 37.0 bits (84), Expect = 0.033
Identities = 31/106 (29%), Positives = 42/106 (39%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G G A+ + G G SSG + S++ +R GSG G ++
Sbjct: 216 GGAGSGAGAGSGSGSGSASTSGSGSGSGSSGAGSEAGSHAGSRAGSGAGSGAG-----SE 380
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRG 862
AG G G G G G + S + GS G S+R S G
Sbjct: 381 AGSYAGSHAGSGSSGAGSEAGS--EAGSYAGSHAGSGSSRAGSEAG 512
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,760,727
Number of Sequences: 36976
Number of extensions: 274457
Number of successful extensions: 3217
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 2308
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2740
length of query: 865
length of database: 9,014,727
effective HSP length: 104
effective length of query: 761
effective length of database: 5,169,223
effective search space: 3933778703
effective search space used: 3933778703
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137602.5


