

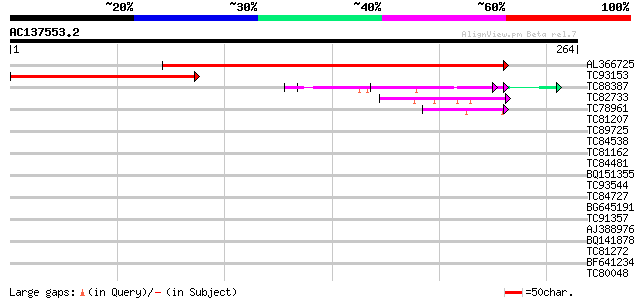
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.2 - phase: 0
(264 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AL366725 255 1e-68
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 169 1e-42
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 48 3e-06
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 46 1e-05
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 42 2e-04
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 40 0.001
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 32 0.33
TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to p... 30 0.95
TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein... 30 0.95
TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis t... 30 1.2
BQ151355 homologue to GP|12620658|gb| ID556 {Bradyrhizobium japo... 30 1.2
TC93544 weakly similar to SP|O82256|COLA_ARATH Zinc finger prote... 30 1.2
TC84727 similar to GP|16118856|gb|AAL14629.1 growth-on protein G... 29 1.6
BG645191 similar to GP|2194137|gb| ESTs gb|R29947 gb|H76702 come... 29 1.6
TC91357 similar to GP|2194137|gb|AAB61112.1| ESTs gb|R29947 gb|H... 29 1.6
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 29 2.1
BQ141878 similar to GP|12697907|db KIAA1681 protein {Homo sapien... 29 2.1
TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 ... 29 2.1
BF641234 similar to GP|20146108|dbj protein phosphatase 2C {Mese... 29 2.1
TC80048 similar to GP|20146108|dbj|BAB88943. protein phosphatase... 29 2.1
>AL366725
Length = 485
Score = 255 bits (651), Expect = 1e-68
Identities = 116/161 (72%), Positives = 137/161 (85%)
Frame = +2
Query: 72 KFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEEDTKAHYKIR 131
KFYPHY+AETAEFSKCIKFENGLR DIKRAIGYQ++R F +LV++ IYEEDTKAH K+
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 132 NERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEE 191
NER+ KGQ RPKPYSAP +KG+QR+ D+RRPK++D PAEIVC+ GEKGHKSNVC E
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEI 361
Query: 192 KKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
KKC RC KK H +A+CKR DIVCFNC+EEGH+ SQCK+PK+
Sbjct: 362 KKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 169 bits (428), Expect = 1e-42
Identities = 81/88 (92%), Positives = 83/88 (94%)
Frame = +2
Query: 1 MLAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
MLAEEADDWW SLLPVLEQD VVTWA+FR+EFL RYFPEDVRGKKEIEFLELKQGDMSV
Sbjct: 251 MLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIEFLELKQGDMSV 430
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCI 88
TEYAAKFVELA FYPHYSAETAEFSKCI
Sbjct: 431 TEYAAKFVELATFYPHYSAETAEFSKCI 514
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 48.1 bits (113), Expect = 3e-06
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 6/65 (9%)
Frame = +1
Query: 169 PAEIVCYKCGEKGHKSNVCN------GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
P E +C+ CG+ GH++ C G+ + C C K+ H EC + C NC + GH
Sbjct: 580 PNEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC-TNEKACNNCRKTGH 756
Query: 223 LTSQC 227
L C
Sbjct: 757 LARDC 771
Score = 46.6 bits (109), Expect = 1e-05
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 5/109 (4%)
Frame = +1
Query: 129 KIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKR-----RDDPAEIVCYKCGEKGHK 183
+IR+ER + R PY +G + N + KR R+ P VC+ C GH
Sbjct: 343 RIRSERFSH----REAPYRRDSRRGFSQDNLCKNCKRPGHYVRECPNVAVCHNCSLPGHI 510
Query: 184 SNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
++ C+ + C+ C + H + C I C C + GH +C P+K
Sbjct: 511 ASECS-TKSLCWNCKEPGHMASSCPNEGI-CHTCGKAGHRARECTVPQK 651
Score = 45.4 bits (106), Expect = 2e-05
Identities = 34/132 (25%), Positives = 50/132 (37%), Gaps = 9/132 (6%)
Frame = +1
Query: 135 RNKGQQGRPKPYSAPVNKGEQRLNDER---------RPKRRDDPAEIVCYKCGEKGHKSN 185
R+K + P S + ++R+ ER R RR + +C C GH
Sbjct: 280 RSKSRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGHYVR 459
Query: 186 VCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDSDYVLT 245
C C C H +EC + C+NC E GH+ S C P +G
Sbjct: 460 ECPNVAV-CHNCSLPGHIASECSTKSL-CWNCKEPGHMASSC--PNEG------------ 591
Query: 246 KVMNKCGELGIR 257
+ + CG+ G R
Sbjct: 592 -ICHTCGKAGHR 624
Score = 33.1 bits (74), Expect = 0.11
Identities = 16/44 (36%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Frame = +1
Query: 171 EIVCYKCGEKGHKSNVC-NGEEKKCFRCGKKWHTIAECKRGDIV 213
++VC C + GH S C G C CG + H EC G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
Score = 30.8 bits (68), Expect = 0.56
Identities = 22/85 (25%), Positives = 30/85 (34%), Gaps = 23/85 (27%)
Frame = +1
Query: 166 RDDPAEIVCYKCGEKGH------KSNVCNGE----------------EKKCFRCGKKWHT 203
RD P + +C C GH KSNV + C C + H
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 204 IAECKRGDI-VCFNCDEEGHLTSQC 227
+C G + +C NC GH +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 29.6 bits (65), Expect = 1.2
Identities = 17/79 (21%), Positives = 27/79 (33%), Gaps = 22/79 (27%)
Frame = +1
Query: 171 EIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRG-------------------- 210
E C C + GH + C + C C H +C +
Sbjct: 721 EKACNNCRKTGHLARDCPNDPI-CNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGG 897
Query: 211 --DIVCFNCDEEGHLTSQC 227
D+VC +C + GH++ C
Sbjct: 898 FRDVVCRSCQQFGHMSRDC 954
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 46.2 bits (108), Expect = 1e-05
Identities = 24/73 (32%), Positives = 38/73 (51%), Gaps = 12/73 (16%)
Frame = +3
Query: 173 VCYKCGEKGHKSNVC-NGEEKKCFR----CGKKWHTIAEC----KRGDIV---CFNCDEE 220
+C +C +GH++ C +G K+ F+ CG H++A C + G + CF C E+
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 221 GHLTSQCKKPKKG 233
GHL+ C K G
Sbjct: 534 GHLSKNCPKNAHG 572
Score = 34.7 bits (78), Expect = 0.039
Identities = 29/156 (18%), Positives = 56/156 (35%), Gaps = 22/156 (14%)
Frame = +3
Query: 110 FSELVSSYMIYEEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDER-----RPK 164
F S+ M+ + A + + E + P P + P K + + ++ +P+
Sbjct: 54 FQSNQSAKMVSQRQRLAKKRYKEEHPELFPKVDPTPPNDPSKKKKNKFKRKKPDSNSKPR 233
Query: 165 RRDDPAEIV-------CYKCGEKGHKSNVCNGE-----EKKCFRCGKKWHTIAECKRGDI 212
P + C+ C H + C + K C RC ++ H C G
Sbjct: 234 TGKRPLRVPGMKPGDSCFICKGLDHIAKFCTQKAEWEKNKICLRCRRRGHRAQNCPDGGS 413
Query: 213 -----VCFNCDEEGHLTSQCKKPKKGQASVYDSDYV 243
+NC + GH + C P + +++ +V
Sbjct: 414 KEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFV 521
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 42.4 bits (98), Expect = 2e-04
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +2
Query: 193 KCFRCGKKWHTIAECKRGD--IVCFNCDEEGHLTSQCK-KPKK 232
+CF CG H +CK GD C+ C E GH+ CK PKK
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKK 484
Score = 37.7 bits (86), Expect = 0.005
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 3/66 (4%)
Frame = +2
Query: 146 YSAPVNKGEQRLNDERRPKRRDDP-AEIVCYKCGEKGHKSNVCNGEE--KKCFRCGKKWH 202
++ V +G + D R R P C+ CG GH + C + KC+RCG++ H
Sbjct: 272 FAKGVPRGSRDSRDSREYLGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGH 451
Query: 203 TIAECK 208
CK
Sbjct: 452 IEKNCK 469
Score = 30.8 bits (68), Expect = 0.56
Identities = 11/23 (47%), Positives = 13/23 (55%)
Frame = +2
Query: 174 CYKCGEKGHKSNVCNGEEKKCFR 196
CY+CGE+GH C KK R
Sbjct: 425 CYRCGERGHIEKNCKNSPKKLSR 493
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 39.7 bits (91), Expect = 0.001
Identities = 21/91 (23%), Positives = 30/91 (32%), Gaps = 31/91 (34%)
Frame = +3
Query: 174 CYKCGEKGHKSNVCN-----------------GEEKKCFRCGKKWHTIAECKR------- 209
CY CG+ GH + C+ ++ C+ CG H +C R
Sbjct: 351 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 530
Query: 210 -------GDIVCFNCDEEGHLTSQCKKPKKG 233
G C+ C GH+ C P G
Sbjct: 531 GGGGYGGGGTSCYRCGGVGHIARDCATPSSG 623
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 31.6 bits (70), Expect = 0.33
Identities = 15/57 (26%), Positives = 19/57 (33%), Gaps = 20/57 (35%)
Frame = +1
Query: 174 CYKCGEKGHKSNVC--------------------NGEEKKCFRCGKKWHTIAECKRG 210
CY CGE GH + C G C+ CG+ H +C G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 26.9 bits (58), Expect = 8.1
Identities = 9/25 (36%), Positives = 12/25 (48%)
Frame = +1
Query: 214 CFNCDEEGHLTSQCKKPKKGQASVY 238
C+NC E GH+ +C G Y
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRY 90
>TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to pre-mRNA
splicing factor ATP-dependent RNA helicase, partial
(11%)
Length = 973
Score = 30.0 bits (66), Expect = 0.95
Identities = 24/78 (30%), Positives = 33/78 (41%), Gaps = 7/78 (8%)
Frame = +2
Query: 157 LNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEE--KKCFRCGKKWHTI-----AECKR 209
L+ ER + +DDP + C G+E K CF CG+ I ECK
Sbjct: 683 LSCERYRELKDDPDSSL----------KEWCKGKEQVKSCFACGQIIEKIDGCNHVECKC 832
Query: 210 GDIVCFNCDEEGHLTSQC 227
G VC+ C E + +C
Sbjct: 833 GKHVCWVCLEIFTSSDEC 886
>TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein T26I12.220
- Arabidopsis thaliana, partial (9%)
Length = 756
Score = 30.0 bits (66), Expect = 0.95
Identities = 10/29 (34%), Positives = 19/29 (65%)
Frame = +2
Query: 203 TIAECKRGDIVCFNCDEEGHLTSQCKKPK 231
++A KR + +C+ C ++GH S+C P+
Sbjct: 389 SVASGKRKNRMCYGCRQKGHNLSECPNPQ 475
>TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis thaliana},
partial (23%)
Length = 727
Score = 29.6 bits (65), Expect = 1.2
Identities = 9/20 (45%), Positives = 15/20 (75%)
Frame = +2
Query: 189 GEEKKCFRCGKKWHTIAECK 208
G+++KC+ CG+ H AEC+
Sbjct: 194 GQQEKCYMCGQVGHLAAECR 253
>BQ151355 homologue to GP|12620658|gb| ID556 {Bradyrhizobium japonicum},
partial (4%)
Length = 1420
Score = 29.6 bits (65), Expect = 1.2
Identities = 12/37 (32%), Positives = 21/37 (56%)
Frame = +2
Query: 131 RNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRD 167
R + +N+G +GR P + N+ R DE+R ++ D
Sbjct: 317 RKQPKNRGGEGRHSPTTKAENQEPPRTKDEKRKEKED 427
>TC93544 weakly similar to SP|O82256|COLA_ARATH Zinc finger protein
constans-like 10. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (45%)
Length = 731
Score = 29.6 bits (65), Expect = 1.2
Identities = 17/60 (28%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Frame = +2
Query: 166 RDDPAEIVCYKCGEKGHKSNVCNGEEKK---CFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
R D A++ C+ C + H +N + + C C TI + C NCD E H
Sbjct: 104 RADSAKL-CFSCDREVHSTNQLFSKHTRSLICDSCDDSPATILCSTESSVFCQNCDWENH 280
>TC84727 similar to GP|16118856|gb|AAL14629.1 growth-on protein GRO10
{Euphorbia esula}, partial (47%)
Length = 936
Score = 29.3 bits (64), Expect = 1.6
Identities = 10/22 (45%), Positives = 13/22 (58%)
Frame = +1
Query: 196 RCGKKWHTIAECKRGDIVCFNC 217
RCG++W ECK CF+C
Sbjct: 778 RCGRQWSCKYECKSCSSFCFSC 843
>BG645191 similar to GP|2194137|gb| ESTs gb|R29947 gb|H76702 come from this
gene. {Arabidopsis thaliana}, partial (40%)
Length = 649
Score = 29.3 bits (64), Expect = 1.6
Identities = 12/21 (57%), Positives = 13/21 (61%), Gaps = 2/21 (9%)
Frame = +1
Query: 174 CYKCGEKGHKSNVCN--GEEK 192
C +CG GH S CN GEEK
Sbjct: 121 CSQCGNNGHNSRTCNDGGEEK 183
>TC91357 similar to GP|2194137|gb|AAB61112.1| ESTs gb|R29947 gb|H76702 come
from this gene. {Arabidopsis thaliana}, partial (40%)
Length = 766
Score = 29.3 bits (64), Expect = 1.6
Identities = 12/21 (57%), Positives = 13/21 (61%), Gaps = 2/21 (9%)
Frame = +3
Query: 174 CYKCGEKGHKSNVCN--GEEK 192
C +CG GH S CN GEEK
Sbjct: 228 CSQCGNNGHNSRTCNDGGEEK 290
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 28.9 bits (63), Expect = 2.1
Identities = 9/19 (47%), Positives = 13/19 (68%)
Frame = +2
Query: 170 AEIVCYKCGEKGHKSNVCN 188
+++ CY CGE GH + CN
Sbjct: 332 SDLKCYXCGEPGHFARXCN 388
>BQ141878 similar to GP|12697907|db KIAA1681 protein {Homo sapiens}, partial
(2%)
Length = 1102
Score = 28.9 bits (63), Expect = 2.1
Identities = 13/45 (28%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Frame = -2
Query: 110 FSELVSSYMIYEEDTKAHYKIRNERRNKGQQGRPKP---YSAPVN 151
FS+L+ + I D K +RN + ++G + P P +++P N
Sbjct: 231 FSDLMMNEYIISNDAKERKNVRNRKIHRGDKNHPNPSLTHTSPPN 97
>TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 -
Arabidopsis thaliana, partial (3%)
Length = 757
Score = 28.9 bits (63), Expect = 2.1
Identities = 20/84 (23%), Positives = 29/84 (33%)
Frame = +2
Query: 134 RRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEKK 193
RR ++G P+ AP ++ + RP DP H+ C+ +
Sbjct: 44 RRQALREGLPQ---APASR-----HSRPRPPAHPDP------------HRPRWCSPARPR 163
Query: 194 CFRCGKKWHTIAECKRGDIVCFNC 217
C RC W C CF C
Sbjct: 164 CQRCSSAWPWCFRCSSARPWCFRC 235
>BF641234 similar to GP|20146108|dbj protein phosphatase 2C {Mesembryanthemum
crystallinum}, partial (13%)
Length = 591
Score = 28.9 bits (63), Expect = 2.1
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = -3
Query: 89 KFENGLRADIKRAIGYQK 106
KFENG D K+ +GYQK
Sbjct: 223 KFENGFNPDCKKELGYQK 170
>TC80048 similar to GP|20146108|dbj|BAB88943. protein phosphatase 2C
{Mesembryanthemum crystallinum}, partial (15%)
Length = 705
Score = 28.9 bits (63), Expect = 2.1
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = -1
Query: 89 KFENGLRADIKRAIGYQK 106
KFENG D K+ +GYQK
Sbjct: 270 KFENGFNPDCKKELGYQK 217
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,716,285
Number of Sequences: 36976
Number of extensions: 131045
Number of successful extensions: 724
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 661
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 702
length of query: 264
length of database: 9,014,727
effective HSP length: 94
effective length of query: 170
effective length of database: 5,538,983
effective search space: 941627110
effective search space used: 941627110
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC137553.2


