

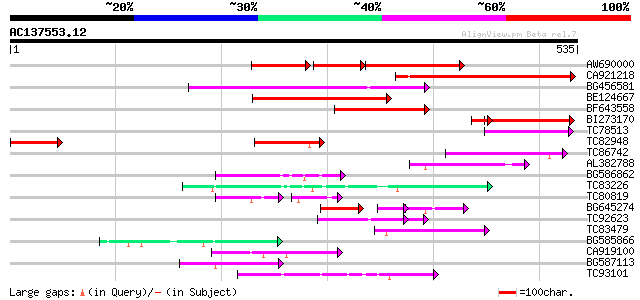
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.12 + phase: 0
(535 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW690000 129 2e-61
CA921218 197 6e-51
BG456581 151 7e-37
BE124667 135 3e-32
BF643558 124 9e-29
BI273170 110 8e-28
TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical prot... 69 6e-12
TC82948 69 6e-12
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 66 3e-11
AL382788 weakly similar to GP|11322386|emb glycine receptor beta... 64 2e-10
BG586862 63 2e-10
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 63 3e-10
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 57 5e-10
BG645274 35 7e-09
TC92623 51 1e-08
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 56 4e-08
BG585866 55 5e-08
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 54 2e-07
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 52 4e-07
TC93101 50 2e-06
>AW690000
Length = 652
Score = 129 bits (323), Expect(3) = 2e-61
Identities = 59/94 (62%), Positives = 76/94 (80%)
Frame = +3
Query: 336 LSRKVASPSISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTDGSTSGSLSSCG 395
LS VAS SISNF+ILKKF VT+HPP+APKIIEVIW+PP +W KCNTDGS+ S+CG
Sbjct: 306 LSNAVASASISNFLILKKFNVTIHPPKAPKIIEVIWRPPIPHWIKCNTDGSSRSHSSACG 485
Query: 396 GIFRNCNSEMLICFGENTGIGNSLHAELSGAMRA 429
GIFRN ++++L+CF ENTG N+ HAEL ++++
Sbjct: 486 GIFRNHDTDLLLCFAENTGECNAFHAELLXSLKS 587
Score = 85.1 bits (209), Expect(3) = 2e-61
Identities = 33/56 (58%), Positives = 45/56 (79%)
Frame = +2
Query: 229 LPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQSKEDLWNICNGSW 284
LPSMCSLC +E++ HLF ECS+A+NLW WFA +LN +++ QS ED+W++CN SW
Sbjct: 2 LPSMCSLCCKQAESSLHLFFECSYAVNLWCWFASVLNKTLHFQSLEDMWSLCNRSW 169
Score = 61.6 bits (148), Expect(3) = 2e-61
Identities = 27/49 (55%), Positives = 38/49 (77%)
Frame = +1
Query: 287 QCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGN 335
Q VI A++INI+NS+W+ARN RF N KIHW+SS++++ISN +L N
Sbjct: 160 QIMEVITATMINIMNSVWFARN*LRFSNKKIHWKSSLSTVISNTALFDN 306
>CA921218
Length = 707
Score = 197 bits (502), Expect = 6e-51
Identities = 91/170 (53%), Positives = 123/170 (71%)
Frame = -3
Query: 365 KIIEVIWQPPPLNWTKCNTDGSTSGSLSSCGGIFRNCNSEMLICFGENTGIGNSLHAELS 424
KI + P PL W CNT+GS + + S+CGG FRN N+ L+CF ENTG GN+LHA+LS
Sbjct: 618 KIESFLALPEPL-WINCNTNGSANINTSACGGTFRNSNAYFLLCFAENTGNGNALHAKLS 442
Query: 425 GAMRAIELAISHQWLNLWLEVDSELVIRAFKNQSIVPWHLRNRWMNCMLLTRNMNFIATH 484
GAMRAIE+A + W +LWLE+DS LV+ AFK++S++ W+LRNRW NC+ L +MNF +H
Sbjct: 441 GAMRAIEIAAARNWSHLWLELDSSLVVNAFKSKSVIHWNLRNRWNNCLFLISSMNFFVSH 262
Query: 485 IFRERNVCADLLAFTGHNLDNFTVWMNVPTCITEPYIKNRLGMPNFRFVT 534
+FRE N CAD LA G +LD+ T W +VP I Y++N++G P FRF++
Sbjct: 261 VFREGNQCADGLANFGLSLDHLTYWNHVPPFIHRFYVENKIGWPMFRFIS 112
>BG456581
Length = 683
Score = 151 bits (381), Expect = 7e-37
Identities = 76/228 (33%), Positives = 123/228 (53%)
Frame = +2
Query: 169 NNGELTLQDAYKFKKTNFPKVNWAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCN 228
+ G+LT++ Y F ++ WA IW+ IPPS + + WR ++LPTDDNL ++GC
Sbjct: 2 HQGKLTIKLVYSFLTSHTSCAPWASTIWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCA 181
Query: 229 LPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQSKEDLWNICNGSWNPQC 288
L SMCS C ET+ HLFL C F + LWSW L ++ S + L + + Q
Sbjct: 182 LVSMCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLRVGLDFSSFKALLSSLPRHCSSQV 361
Query: 289 KTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSISNF 348
+ + +A+++++++SIW+ARN RF + K+ + + + L+G +S ++
Sbjct: 362 RDLYVAAVVHMVHSIWWARNNVRFSSAKVSAHAVQVRVHALIGLSGAVS--TGKCIAADA 535
Query: 349 VILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTDGSTSGSLSSCGG 396
IL F + H +++ V W+PP W K NTDGS + + GG
Sbjct: 536 AILDVFRIPPHRRSMXEMVSVCWKPPSAPWVKGNTDGSXLNNSGAXGG 679
>BE124667
Length = 540
Score = 135 bits (341), Expect = 3e-32
Identities = 60/131 (45%), Positives = 85/131 (64%)
Frame = -1
Query: 230 PSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQSKEDLWNICNGSWNPQCK 289
PSMCS C SETT HLF EC+FA +WSW A +LN + + D+W+ +W PQCK
Sbjct: 393 PSMCSSCQASSETTFHLFFECNFATKMWSWLASLLNTPLQFSTSADMWSPLQLNWTPQCK 214
Query: 290 TVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSISNFV 349
VI A +IN++N IW+ RN RFQ+ I W+++I IIS SL+GNL++K ++ ++ F
Sbjct: 213 VVITACIINLLNVIWFRRNNIRFQDKVIDWKTAINMIISKVSLSGNLTKKTSAANMLEFT 34
Query: 350 ILKKFCVTLHP 360
I K V++ P
Sbjct: 33 IFKACKVSIKP 1
>BF643558
Length = 645
Score = 124 bits (311), Expect = 9e-29
Identities = 55/90 (61%), Positives = 70/90 (77%)
Frame = +2
Query: 307 RNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSISNFVILKKFCVTLHPPRAPKI 366
RNQRRF N IHW+SSI SIIS+ L+GN + +I++FV LKKF + +HPP+APKI
Sbjct: 365 RNQRRFNNKMIHWKSSITSIISSTFLSGNYTSACDYTNITDFVFLKKFSIDIHPPKAPKI 544
Query: 367 IEVIWQPPPLNWTKCNTDGSTSGSLSSCGG 396
IEV+W PPPL+W KCNTDGS++ + SSCGG
Sbjct: 545 IEVLWNPPPLHWIKCNTDGSSNTNTSSCGG 634
>BI273170
Length = 391
Score = 110 bits (274), Expect(2) = 8e-28
Identities = 50/85 (58%), Positives = 62/85 (72%)
Frame = +1
Query: 449 LVIRAFKNQSIVPWHLRNRWMNCMLLTRNMNFIATHIFRERNVCADLLAFTGHNLDNFTV 508
L+ FKN S VPW LRNRW NC+L TRNMNFI +H+FRE N CAD+LA G +L+ T+
Sbjct: 52 LLSMPFKNISQVPWKLRNRWENCILATRNMNFIVSHVFREGNECADMLANIGLSLNCLTI 231
Query: 509 WMNVPTCITEPYIKNRLGMPNFRFV 533
W+ +P CI +IKN+LG PN RFV
Sbjct: 232 WLELPDCIKSIFIKNKLGWPNNRFV 306
Score = 32.0 bits (71), Expect(2) = 8e-28
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +3
Query: 436 HQWLNLWLEVDSELVIRAFK 455
H W +LWLE DS LV+ AF+
Sbjct: 12 HNWQSLWLESDSALVVNAFQ 71
>TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical protein {Galega
orientalis}, partial (20%)
Length = 765
Score = 68.6 bits (166), Expect = 6e-12
Identities = 33/84 (39%), Positives = 49/84 (58%)
Frame = +2
Query: 449 LVIRAFKNQSIVPWHLRNRWMNCMLLTRNMNFIATHIFRERNVCADLLAFTGHNLDNFTV 508
LV+ K+ + +PW+L NRW N ++ + + I +HI+RE N AD LA G +LD+
Sbjct: 404 LVVLPSKSTNQIPWNL*NRWNNVKVILQGLKCIISHIYREGNQVADTLANHGLSLDSIHF 583
Query: 509 WMNVPTCITEPYIKNRLGMPNFRF 532
W +VP Y +N+ G NFRF
Sbjct: 584 WNDVPEYTRASYFRNKDGWSNFRF 655
>TC82948
Length = 705
Score = 68.6 bits (166), Expect = 6e-12
Identities = 27/69 (39%), Positives = 46/69 (66%), Gaps = 3/69 (4%)
Frame = +3
Query: 232 MCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQSKEDLWNICN---GSWNPQC 288
MCSLC E+THHLF ECS ++ +W WF+ ++N + +I + +DLW IC+ G+ N
Sbjct: 441 MCSLCFKSFESTHHLFFECSVSVQIWDWFSNLINLNFHINNTDDLWRICDREVGAANVSW 620
Query: 289 KTVIIASLI 297
+++++S +
Sbjct: 621 LSLLLSSTL 647
Score = 61.6 bits (148), Expect = 7e-10
Identities = 27/50 (54%), Positives = 35/50 (70%)
Frame = +3
Query: 1 MVTVAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQWAQVLR 50
+V V+W+KVC EG LGIRSL LN A N+K+CWD+ S+EQW +R
Sbjct: 258 VVKVSWEKVCRPIKEGSLGIRSLSKLNEALNLKLCWDMMISKEQWYAFMR 407
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 66.2 bits (160), Expect = 3e-11
Identities = 42/117 (35%), Positives = 56/117 (46%), Gaps = 2/117 (1%)
Frame = -1
Query: 412 NTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVIRAFKNQSIVPWHLRNRWMNC 471
N G L AE M AIE A+ N+ LE DS V+ AF +PW +R RW NC
Sbjct: 423 NIGHATPLEAEFCACMIAIEKAMELGLNNICLETDSLKVVNAFHKIVGIPWQMRVRWHNC 244
Query: 472 MLLTRNMNFIATHIFRERNVCADLLAFTGHNLDNFTV--WMNVPTCITEPYIKNRLG 526
+ ++ + HI RE N+ AD LA G L F + W P+ I ++R G
Sbjct: 243 IRFCHSIACVCVHIPREGNLVADALARHGQGLSLFFLQWWPAPPSFIQSFLAQDRYG 73
>AL382788 weakly similar to GP|11322386|emb glycine receptor betaZ subunit
{Danio rerio}, partial (4%)
Length = 353
Score = 63.5 bits (153), Expect = 2e-10
Identities = 40/115 (34%), Positives = 54/115 (46%), Gaps = 2/115 (1%)
Frame = +2
Query: 378 WTKCNTDGSTSGSL--SSCGGIFRNCNSEMLICFGENTGIGNSLHAELSGAMRAIELAIS 435
W K D +T GS S G +FR+ CF G+ ++HAE A A+E+
Sbjct: 11 WIKAKIDNATCGSSDPSIVGSLFRDQFGSNFGCFIIFFGVNFAIHAEFYAAAYAVEIVYL 190
Query: 436 HQWLNLWLEVDSELVIRAFKNQSIVPWHLRNRWMNCMLLTRNMNFIATHIFRERN 490
W N WL D +LV+ AF + V W L + MNF A HI++ERN
Sbjct: 191 CNWHNFWLACDLKLVVDAFHSNYKVLWSLAT*F-----FCNQMNF*AFHIYKERN 340
>BG586862
Length = 804
Score = 63.2 bits (152), Expect = 2e-10
Identities = 42/127 (33%), Positives = 57/127 (44%), Gaps = 4/127 (3%)
Frame = -1
Query: 195 IWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFAL 254
+W P +WR + N LP D L +G +C C + ET HLFL C
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 255 NLWSWFAGILNCSINIQSKEDL----WNICNGSWNPQCKTVIIASLINIINSIWYARNQR 310
WF L IN S L W I N +T+I +L ++ SIW+ARNQ+
Sbjct: 474 K--EWFGSQL--GINFHSSGVLHFHDW-ITNFILKNDEETII--ALTALLYSIWHARNQK 316
Query: 311 RFQNIKI 317
F+NI +
Sbjct: 315 VFENIDV 295
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 62.8 bits (151), Expect = 3e-10
Identities = 75/313 (23%), Positives = 124/313 (38%), Gaps = 21/313 (6%)
Frame = +3
Query: 164 IWKHSNNGELTLQDAYKFKKT-NFPKVN-----------WAKHIWSPDIPPSKALLVWRF 211
+W H+ G +++ Y +T ++N W K IWS P +L+WR
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKK-IWSLHTIPRHKVLLWRI 179
Query: 212 MLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQ 271
+ + LP +L +G +C C++ +ET HLF+ C + + WF N IN
Sbjct: 180 LNDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRV--WFGS--NLCINFD 347
Query: 272 SKEDLWNICNGSW-----NPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASI 326
+ + N N + +C T+ IA+ II ++W+ARN ++ I I
Sbjct: 348 NLPNP-NFIN*LYEAIL*KDECITI*IAA---IIYNLWHARNLSVLEDQTI-LEMDIIQR 512
Query: 327 ISNASLTGNLSRKVASPSISNFVILKKFCVTLHPPRAP--KIIEVIWQPPPLNWTKCNTD 384
SN + A PS++ T + PR+ W+ P L K NTD
Sbjct: 513 ASNCISDYKQANTQAPPSMAR---------TGYDPRSQHRPAKNTKWKRPNLGLVKVNTD 665
Query: 385 GSTSG-SLSSCGGIFRN-CNSEMLICFGENTGIGNSLHAELSGAMRAIELAISHQWLNLW 442
+ G I R+ M E G +L AE + + A + +
Sbjct: 666 ANLQNHGKWGLGIIIRDEVGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVX 845
Query: 443 LEVDSELVIRAFK 455
E D+E +++ K
Sbjct: 846 FEGDNEKLMKMVK 884
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 57.4 bits (137), Expect(2) = 5e-10
Identities = 28/68 (41%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Frame = +2
Query: 195 IWSPDIPPSKALLVWRFMLNKLPTDDNLMNKG----CNLPSMCSLCNNMSETTHHLFLEC 250
+W IP +L VWR + N+LPT DNL+++G N +C + SE+T HLFL C
Sbjct: 626 VWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATNAACVCGCVD--SESTTHLFLHC 799
Query: 251 SFALNLWS 258
+ +LWS
Sbjct: 800 NVFCSLWS 823
Score = 24.6 bits (52), Expect(2) = 5e-10
Identities = 14/50 (28%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Frame = +1
Query: 267 SINIQ--SKEDLWNICNGSWNPQCKTVIIASLINIINSIWYARNQRRFQN 314
SIN+ + +++C W+ + + I+ +I W RN R FQN
Sbjct: 847 SINVFW*ASNTFYSVCQNGWHAKSFSFILQVNFWVI---WKERNNRLFQN 987
>BG645274
Length = 641
Score = 35.4 bits (80), Expect(3) = 7e-09
Identities = 23/62 (37%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Frame = -3
Query: 374 PPLNWTKCNTDGSTSGSL--SSCGGIFRNCNSEMLICFGENTGIGNSLHAELSGAMRAIE 431
P L+W KCN DGS G ++C GIFR + + I N S AE M IE
Sbjct: 369 PILSWVKCNIDGSIKGCFRPATCSGIFR--DEDA*I*VAVNMHNTPSFEAEFHDLMYEIE 196
Query: 432 LA 433
++
Sbjct: 195 IS 190
Score = 32.3 bits (72), Expect(3) = 7e-09
Identities = 14/41 (34%), Positives = 26/41 (63%)
Frame = -2
Query: 294 ASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTG 334
+++IN+IN IW RN++ F K++ +++ SN L+G
Sbjct: 607 SAIINVINIIW*CRNKQLFNGFKVNSKTTANLGCSNTLLSG 485
Score = 29.3 bits (64), Expect(3) = 7e-09
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = -1
Query: 348 FVILKKFCVTLHPPRAPKIIEVIWQPPPL 376
F IL+ F V H P A +I V+W PP L
Sbjct: 446 FEILRVFFV*DHAPNASRIKAVLWYPPSL 360
>TC92623
Length = 549
Score = 51.2 bits (121), Expect(2) = 1e-08
Identities = 28/86 (32%), Positives = 49/86 (56%)
Frame = +1
Query: 291 VIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSISNFVI 350
V A+L++ +++IW ARN RF + + +S+A I S +L+G S+ + +++ +
Sbjct: 52 VFAAALVHTVHTIWLARNAIRFSSANVSIHTSMAKISSLVALSGANSK--GNCLVTDGAV 225
Query: 351 LKKFCVTLHPPRAPKIIEVIWQPPPL 376
L F + R +II VIW+P PL
Sbjct: 226 LNNFMIPPSYRRVKEIISVIWKPQPL 303
Score = 26.2 bits (56), Expect(2) = 1e-08
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = +3
Query: 372 QPPPLNWTKCNTDGSTSGSLSSCG 395
+ P + W K NTDGS SS G
Sbjct: 288 ETPTITWVKANTDGSVLNLHSSRG 359
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 55.8 bits (133), Expect = 4e-08
Identities = 32/110 (29%), Positives = 52/110 (47%), Gaps = 2/110 (1%)
Frame = +2
Query: 345 ISNFVILKKF--CVTLHPPRAPKIIEVIWQPPPLNWTKCNTDGSTSGSLSSCGGIFRNCN 402
I V+LK + P + II W+ P + WTK NTDGS + + GG+ R+
Sbjct: 893 IKGMVLLKNLNQIANILNPVSRSIIWCEWKKPEIGWTKLNTDGSVNKETAGFGGLLRDYR 1072
Query: 403 SEMLICFGENTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVIR 452
E + F G++ EL R + L++ ++W+E DS V++
Sbjct: 1073 GEPICAFVSKAPQGDTFLVELWAIWRGLVLSLGLGIKSIWVESDSMSVVK 1222
>BG585866
Length = 828
Score = 55.5 bits (132), Expect = 5e-08
Identities = 43/184 (23%), Positives = 74/184 (39%), Gaps = 11/184 (5%)
Frame = +3
Query: 85 WLVGDGDTINCWLDNWCGETLVDLFD--IDSQQLNMLPKKL----RNYMQNFNWCFPDDI 138
W G G++ + W NW L+ +D L++ K + + Q+ P DI
Sbjct: 123 WRAGSGNS-SFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQHTQSLYTILPTDI 299
Query: 139 LSLFPDMRLLASKVTIPKHCIRDKLIWKHSNNGELTLQDAYKF-----KKTNFPKVNWAK 193
+ + L + I D IW H++NG T + Y + + N+ +W+
Sbjct: 300 AEVINNTHLNFNA------SIGDAYIWPHNSNGVYTAKSGYSWILSQTETVNYNNSSWS- 458
Query: 194 HIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFA 253
IW IP +W N +PT L ++ ++CS C E+ H +C F+
Sbjct: 459 WIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVRDCRFS 638
Query: 254 LNLW 257
+W
Sbjct: 639 KIIW 650
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 53.5 bits (127), Expect = 2e-07
Identities = 33/130 (25%), Positives = 61/130 (46%), Gaps = 6/130 (4%)
Frame = -2
Query: 191 WAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNN---MSETTHHLF 247
+A+ IW +P ++ WR + ++LPT NL+ +G +P+ LC + E+ HLF
Sbjct: 596 YAEMIWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGV-IPTEAGLCVSGCGALESAQHLF 420
Query: 248 LECSFALNLWSW---FAGILNCSINIQSKEDLWNICNGSWNPQCKTVIIASLINIINSIW 304
L CS+ +LWS + G + N+ S + + + N ++ + + +W
Sbjct: 419 LSCSYFASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAWVLW 240
Query: 305 YARNQRRFQN 314
RN F +
Sbjct: 239 TERNNMCFND 210
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 52.4 bits (124), Expect = 4e-07
Identities = 25/110 (22%), Positives = 47/110 (42%), Gaps = 12/110 (10%)
Frame = -3
Query: 161 DKLIWKHSNNGELTLQDAYKFKKTNFPKVNWA------------KHIWSPDIPPSKALLV 208
D W++S +G +++ Y + N + +W + P +
Sbjct: 426 DSYSWEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFL 247
Query: 209 WRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWS 258
WR + N LPT N+ ++ + CS C SET +H+ +C +A +W+
Sbjct: 246 WRCISNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIWA 97
>TC93101
Length = 675
Score = 50.4 bits (119), Expect = 2e-06
Identities = 50/192 (26%), Positives = 77/192 (40%), Gaps = 3/192 (1%)
Frame = -1
Query: 216 LPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQSKED 275
LP L +G N P +C C ETT+H+F+ C + WF L+ S +
Sbjct: 663 LPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRV--WFGSQLSIRFPDNSTIN 490
Query: 276 LWNICNGSWNPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGN 335
+ + + Q + +II + I SIW+ARN+ F+N + +I N+ L
Sbjct: 489 FSDWLFDAISNQTEEIII-KISAITYSIWHARNKAIFENQFVS-EDTIIQ*AQNSILA-- 322
Query: 336 LSRKVASPSISNFVILKKFCVT--LHPPRAPKIIEVIWQPPPLNWTKCNTDGSTS-GSLS 392
+ P N ++L T + R + WQ P N K N D +
Sbjct: 321 YEQATIKPQNPN-IVLSSLSATSNTNTTRRRSNVRSRWQKPLNNILKANCDANLQVQGRW 145
Query: 393 SCGGIFRNCNSE 404
G I RN + E
Sbjct: 144 GLGCIIRNADGE 109
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.136 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,335,248
Number of Sequences: 36976
Number of extensions: 452088
Number of successful extensions: 3325
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 3173
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3302
length of query: 535
length of database: 9,014,727
effective HSP length: 101
effective length of query: 434
effective length of database: 5,280,151
effective search space: 2291585534
effective search space used: 2291585534
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137553.12


