

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.2 - phase: 0
(623 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
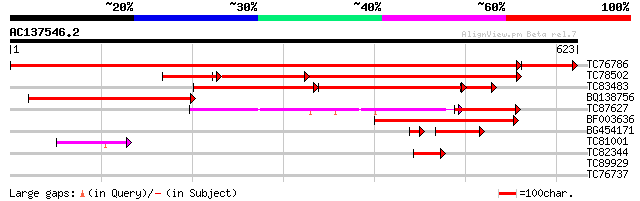
Score E
Sequences producing significant alignments: (bits) Value
TC76786 similar to SP|P51615|MAOX_VITVI NADP-dependent malic enz... 1114 0.0
TC78502 similar to GP|8118507|gb|AAF73006.1| NADP-dependent mali... 386 e-179
TC83483 homologue to GP|510876|emb|CAA56354.1| NADP dependent ma... 288 e-153
BQ138756 homologue to GP|510876|emb| NADP dependent malic enzyme... 335 2e-92
TC87627 similar to SP|P37221|MAOM_SOLTU NAD-dependent malic enzy... 219 3e-57
BF003636 similar to GP|8894554|emb NAD-dependent malic enzyme (m... 136 2e-32
BG454171 homologue to GP|8894554|emb| NAD-dependent malic enzyme... 59 6e-11
TC81001 similar to SP|P37225|MAON_SOLTU NAD-dependent malic enzy... 62 7e-10
TC82344 similar to SP|P37221|MAOM_SOLTU NAD-dependent malic enzy... 42 5e-04
TC89929 similar to GP|10187185|emb|CAC09066. cDNA~Lemon acyl tra... 29 6.2
TC76737 homologue to SP|P06379|RK2_TOBAC Chloroplast 50S ribosom... 28 8.0
>TC76786 similar to SP|P51615|MAOX_VITVI NADP-dependent malic enzyme (EC
1.1.1.40) (NADP-ME). [Grape] {Vitis vinifera}, complete
Length = 2039
Score = 1114 bits (2881), Expect(2) = 0.0
Identities = 562/562 (100%), Positives = 562/562 (100%)
Frame = +1
Query: 1 MESTLKSFRDGESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQ 60
MESTLKSFRDGESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQ
Sbjct: 100 MESTLKSFRDGESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQ 279
Query: 61 YNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNE 120
YNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNE
Sbjct: 280 YNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNE 459
Query: 121 RLFYKLLIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPER 180
RLFYKLLIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPER
Sbjct: 460 RLFYKLLIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPER 639
Query: 181 SIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLL 240
SIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLL
Sbjct: 640 SIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLL 819
Query: 241 NDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTH 300
NDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTH
Sbjct: 820 NDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTH 999
Query: 301 LVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKA 360
LVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKA
Sbjct: 1000LVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKA 1179
Query: 361 PIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSG 420
PIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSG
Sbjct: 1180PIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSG 1359
Query: 421 VGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEY 480
VGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEY
Sbjct: 1360VGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEY 1539
Query: 481 KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPP 540
KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPP
Sbjct: 1540KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPP 1719
Query: 541 FSDIRKISANIAANVAAKAYEL 562
FSDIRKISANIAANVAAKAYEL
Sbjct: 1720FSDIRKISANIAANVAAKAYEL 1785
Score = 129 bits (325), Expect(2) = 0.0
Identities = 60/61 (98%), Positives = 61/61 (99%)
Frame = +2
Query: 563 VCTATIGEQGQFTIDSEAFGRIKDVVGVWCPFMLVKNKRFSALSEPICSFHYDILFHLMS 622
+CTATIGEQGQFTIDSEAFGRIKDVVGVWCPFMLVKNKRFSALSEPICSFHYDILFHLMS
Sbjct: 1853 LCTATIGEQGQFTIDSEAFGRIKDVVGVWCPFMLVKNKRFSALSEPICSFHYDILFHLMS 2032
Query: 623 F 623
F
Sbjct: 2033 F 2035
>TC78502 similar to GP|8118507|gb|AAF73006.1| NADP-dependent malic protein
{Ricinus communis}, partial (65%)
Length = 1540
Score = 386 bits (991), Expect(3) = e-179
Identities = 189/238 (79%), Positives = 212/238 (88%)
Frame = +2
Query: 325 IGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRAN 384
+GG L +H FLFLGAGEAGTGIAELIALE SKQT AP++E RK IWLVDSKGLIVSSR
Sbjct: 473 VGGNLADHKFLFLGAGEAGTGIAELIALETSKQTNAPLDEVRKNIWLVDSKGLIVSSRKE 652
Query: 385 SLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILA 444
SLQHFKKPWAHEHEPV LLDAV IKPTVLIG+SG G FTKEVVEAM IN+ P+IL+
Sbjct: 653 SLQHFKKPWAHEHEPVKNLLDAVNKIKPTVLIGTSGQGSAFTKEVVEAMASINEKPIILS 832
Query: 445 LSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIV 504
LSNPTSQSECTAEEAYTW++GRAIFASGSPF PVEYKGK + GQ+NNAYIFPGFGLG++
Sbjct: 833 LSNPTSQSECTAEEAYTWTQGRAIFASGSPFSPVEYKGKVFVPGQANNAYIFPGFGLGLI 1012
Query: 505 MSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
MSG IRVHDD+LLAASEALA+QV+EEN++KGL +PPF++IRKISA+IAA VAAKAYEL
Sbjct: 1013MSGTIRVHDDLLLAASEALAEQVSEENFEKGLIFPPFTNIRKISAHIAAKVAAKAYEL 1186
Score = 161 bits (408), Expect(3) = e-179
Identities = 80/106 (75%), Positives = 94/106 (88%)
Frame = +1
Query: 224 ACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFED 283
ACL + + GTNNE+LLNDE YIGL+Q+RATG+EY +L+HEFMTAVKQ YGEKVL+QFED
Sbjct: 172 ACLLLLMS-GTNNERLLNDELYIGLKQRRATGQEYSELMHEFMTAVKQTYGEKVLIQFED 348
Query: 284 FANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTL 329
FANHNAF+LL KY +THLVFNDDIQGTA+VVLAG+VA+LKL G L
Sbjct: 349 FANHNAFDLLEKYRSTHLVFNDDIQGTASVVLAGLVAALKLGWGKL 486
Score = 121 bits (303), Expect(3) = e-179
Identities = 56/64 (87%), Positives = 63/64 (97%)
Frame = +3
Query: 169 KILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPV 228
+I EVL+NWPE++IQVIVVTDGERILGLGDLGCQGMGIPVGKL+LY+ALGGVRPSACLP+
Sbjct: 3 RIQEVLRNWPEKNIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACLPI 182
Query: 229 TIDV 232
TIDV
Sbjct: 183 TIDV 194
>TC83483 homologue to GP|510876|emb|CAA56354.1| NADP dependent malic enzyme
{Phaseolus vulgaris}, partial (56%)
Length = 1003
Score = 288 bits (738), Expect(3) = e-153
Identities = 141/163 (86%), Positives = 152/163 (92%)
Frame = +3
Query: 340 GEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEP 399
G+AGTGIAELIALE+SKQTKAP+EE+RKKIWLVDSKGLIVSSR SLQHFKKPWAHEHEP
Sbjct: 414 GKAGTGIAELIALEISKQTKAPVEETRKKIWLVDSKGLIVSSRLQSLQHFKKPWAHEHEP 593
Query: 400 VSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEA 459
V LL+AVK IKPTVLIGSSGVGKTFTKEVVEAM +N+ PLILALSNPTSQSECTAEEA
Sbjct: 594 VKELLNAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEA 773
Query: 460 YTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLG 502
YTWS+G+AIFASGSPFDPVEY+GK + GQSNNAYIFPGFGLG
Sbjct: 774 YTWSKGKAIFASGSPFDPVEYEGKVFVPGQSNNAYIFPGFGLG 902
Score = 242 bits (617), Expect(3) = e-153
Identities = 117/137 (85%), Positives = 130/137 (94%)
Frame = +2
Query: 203 GMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLL 262
GMGIPVGKL+LY+ALGGVRPS+CLP+TIDVGTNNEKLLNDEFYIGL QKRATGKEY +LL
Sbjct: 2 GMGIPVGKLSLYTALGGVRPSSCLPITIDVGTNNEKLLNDEFYIGLXQKRATGKEYAELL 181
Query: 263 HEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASL 322
EFM AVKQNYGEKVLVQFEDFANHNAF+LL KY ++HLVFNDDIQGTA+VVLAG++ASL
Sbjct: 182 EEFMHAVKQNYGEKVLVQFEDFANHNAFDLLDKYSSSHLVFNDDIQGTASVVLAGLLASL 361
Query: 323 KLIGGTLPEHTFLFLGA 339
KL+GGTL +HTFLFLGA
Sbjct: 362 KLVGGTLADHTFLFLGA 412
Score = 51.6 bits (122), Expect(3) = e-153
Identities = 24/39 (61%), Positives = 31/39 (78%)
Frame = +1
Query: 496 FPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKK 534
FP G+++SGAIRV D+MLLA SEALA QV++ENY +
Sbjct: 883 FPDLVWGLIISGAIRVRDEMLLAXSEALAAQVSQENYDR 999
>BQ138756 homologue to GP|510876|emb| NADP dependent malic enzyme {Phaseolus
vulgaris}, partial (30%)
Length = 680
Score = 335 bits (860), Expect = 2e-92
Identities = 157/184 (85%), Positives = 176/184 (95%)
Frame = +2
Query: 21 SAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEKERDAHYLRGL 80
+ VGGGV+D+YGED ATEDQL+TPW FSVASG +LLRDP+YNKGLAFTEKERDAHY+RGL
Sbjct: 128 NGVGGGVKDLYGEDSATEDQLITPWNFSVASGCTLLRDPRYNKGLAFTEKERDAHYVRGL 307
Query: 81 LPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYT 140
LPP V +Q+LQEK+LMHN+RQY+VPL KY+A+MDLQERNERLFYKL+IDNVEELLP+VYT
Sbjct: 308 LPPAVFTQELQEKRLMHNLRQYDVPLHKYIALMDLQERNERLFYKLMIDNVEELLPVVYT 487
Query: 141 PVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLG 200
P VGEACQKYGSIY+RPQGLYISLKEKGKILEVLKNWPE++IQVIVVTDGERILGLGDLG
Sbjct: 488 PTVGEACQKYGSIYRRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLG 667
Query: 201 CQGM 204
CQGM
Sbjct: 668 CQGM 679
>TC87627 similar to SP|P37221|MAOM_SOLTU NAD-dependent malic enzyme 62 kDa
isoform mitochondrial precursor (EC 1.1.1.39) (NAD-ME).
[Potato], partial (67%)
Length = 1548
Score = 219 bits (557), Expect = 3e-57
Identities = 127/316 (40%), Positives = 184/316 (58%), Gaps = 15/316 (4%)
Frame = +2
Query: 198 DLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKE 257
DLG QG+GI +GKL LY A G+ P LPV IDVGTNN+KLL D Y+GL+Q R G +
Sbjct: 2 DLGVQGIGITIGKLDLYVAAAGINPQRVLPVMIDVGTNNKKLLEDPLYLGLQQHRLDGDD 181
Query: 258 YYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAG 317
Y ++ EFM AV + V+VQFEDF + AF+LL +Y TT+ +FNDD+QGTA V +AG
Sbjct: 182 YLAVIDEFMEAVFTRW-PNVIVQFEDFQSKWAFKLLQRYRTTYRMFNDDVQGTAGVAIAG 358
Query: 318 VVASLKLIGGTL---PEHTFLFLGAGEAGTGIAELIALEMSK---QTKAPIEESRKKIWL 371
++ +++ G + P+ + GAG AG G+ M++ + + ++ + W+
Sbjct: 359 LLGAVRAQGRPMIDFPKQKIVVAGAGSAGIGVLNAARKTMARMLGNNEVAFQSAKSQFWV 538
Query: 372 VDSKGLIVSSRANSLQHFKKPWAHEHEPV--------STLLDAVKIIKPTVLIGSSGVGK 423
VD+KGLI R N + P+A + + ++L + VK +KP VL+G S VG
Sbjct: 539 VDAKGLITEGREN-IDPDALPFARNLKEMDRQGLREGASLAEVVKQVKPDVLLGLSAVGG 715
Query: 424 TFTKEVVEAMTEINKI-PLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKG 482
F+ EV+EA+ + P I A+SNPT +ECT +EA++ IFASGSPF V+
Sbjct: 716 LFSNEVLEALKDSTSTRPAIFAMSNPTKNAECTPDEAFSILGDNIIFASGSPFSNVDL-- 889
Query: 483 KTYYSGQSNNAYIFPG 498
G N ++ PG
Sbjct: 890 -----GNGNIGHLQPG 922
Score = 68.6 bits (166), Expect = 7e-12
Identities = 36/73 (49%), Positives = 47/73 (64%)
Frame = +3
Query: 489 QSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKIS 548
Q NN Y+FPG GLG ++SG+ V D ML AA+E LA ++EE KG+ +P S IR I+
Sbjct: 918 QGNNMYLFPGIGLGTLLSGSRIVSDGMLQAAAERLAAYMSEEEVLKGIIFPSISRIRDIT 1097
Query: 549 ANIAANVAAKAYE 561
IAA V +A E
Sbjct: 1098KEIAAAVIEEAVE 1136
>BF003636 similar to GP|8894554|emb NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (66%)
Length = 608
Score = 136 bits (343), Expect = 2e-32
Identities = 71/161 (44%), Positives = 106/161 (65%), Gaps = 2/161 (1%)
Frame = +2
Query: 401 STLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTE-INKIPLILALSNPTSQSECTAEEA 459
+++++ VK + P VL+G SGVG F ++V++AM E ++ P I A+SNPT +ECTA +A
Sbjct: 26 ASIIEVVKKVLPHVLLGLSGVGGVFNEQVLKAMRESVSTKPAIFAMSNPTMNAECTAIDA 205
Query: 460 YTWSEGRAIFASGSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLA 518
+ + +FASGSPF+ V+ G+ + Q+NN Y+FPG GLG ++SGA + D ML A
Sbjct: 206 FNHAGENIVFASGSPFENVDLGNGRAGHVNQANNMYLFPGIGLGSLLSGAHHITDGMLQA 385
Query: 519 ASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKA 559
ASE LA +TEE+ +KG+ YP IR ++A + A V A
Sbjct: 386 ASECLASYMTEEDIQKGILYPSIDCIRNVTAEVGAAVLRAA 508
>BG454171 homologue to GP|8894554|emb| NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (26%)
Length = 487
Score = 58.9 bits (141), Expect(2) = 6e-11
Identities = 29/55 (52%), Positives = 39/55 (70%), Gaps = 1/55 (1%)
Frame = +3
Query: 468 IFASGSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASE 521
+FASGSPF+ V+ G+ + Q+NN Y+FPG GLG ++SGA + D ML AASE
Sbjct: 105 VFASGSPFENVDLGNGRAGHVNQANNMYLFPGIGLGSLLSGAHHITDGMLQAASE 269
Score = 26.6 bits (57), Expect(2) = 6e-11
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +1
Query: 440 PLILALSNPTSQSECT 455
P I A+SNPT +ECT
Sbjct: 19 PAIFAMSNPTMNAECT 66
>TC81001 similar to SP|P37225|MAON_SOLTU NAD-dependent malic enzyme 59 kDa
isoform mitochondrial precursor (EC 1.1.1.39) (NAD-ME).
[Potato], partial (20%)
Length = 679
Score = 62.0 bits (149), Expect = 7e-10
Identities = 36/92 (39%), Positives = 50/92 (54%), Gaps = 10/92 (10%)
Frame = +1
Query: 52 GYSLLRDPQYNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYE-------- 103
G +L DP +NK F ERD LRGLLPP V S + Q + + + R E
Sbjct: 367 GADILHDPWFNKDTGFPLTERDRLGLRGLLPPRVISFEQQYDRFLDSYRSLEKNTLGQPE 546
Query: 104 --VPLQKYVAMMDLQERNERLFYKLLIDNVEE 133
V L K+ + L +RNE L+Y++LIDN++E
Sbjct: 547 NVVSLAKWRILNRLHDRNETLYYRVLIDNIKE 642
>TC82344 similar to SP|P37221|MAOM_SOLTU NAD-dependent malic enzyme 62 kDa
isoform mitochondrial precursor (EC 1.1.1.39) (NAD-ME).
[Potato], partial (5%)
Length = 675
Score = 42.4 bits (98), Expect = 5e-04
Identities = 19/36 (52%), Positives = 25/36 (68%)
Frame = +1
Query: 444 ALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVE 479
A+SNPT +ECT +EA++ IFASGSPF V+
Sbjct: 1 AMSNPTKNAECTPDEAFSILGDNIIFASGSPFSNVD 108
>TC89929 similar to GP|10187185|emb|CAC09066. cDNA~Lemon acyl transferase
{Citrus limon}, partial (22%)
Length = 1361
Score = 28.9 bits (63), Expect = 6.2
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 6/52 (11%)
Frame = +3
Query: 389 FKKPWAH------EHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMT 434
F K WA+ E E + TLL +K + +I +G+G FTK E +T
Sbjct: 459 FIKAWAYLCNKTIETEELPTLLPELKPLLDREIIKDNGLGDKFTKNWTEIIT 614
>TC76737 homologue to SP|P06379|RK2_TOBAC Chloroplast 50S ribosomal protein
L2. [Common tobacco] {Nicotiana tabacum}, partial (95%)
Length = 5466
Score = 28.5 bits (62), Expect = 8.0
Identities = 19/68 (27%), Positives = 32/68 (46%)
Frame = +1
Query: 516 LLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYELVCTATIGEQGQFT 575
L A+ A+AK + +E L P ++R IS N C+AT+G+ G
Sbjct: 1096 LARAAGAVAKLIAKEGKSATLKLPS-GEVRLISKN-------------CSATVGQVGNVG 1233
Query: 576 IDSEAFGR 583
++ ++ GR
Sbjct: 1234 VNQKSLGR 1257
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,242,025
Number of Sequences: 36976
Number of extensions: 220460
Number of successful extensions: 1059
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1048
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1053
length of query: 623
length of database: 9,014,727
effective HSP length: 102
effective length of query: 521
effective length of database: 5,243,175
effective search space: 2731694175
effective search space used: 2731694175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137546.2


