

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.5 - phase: 0
(1519 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
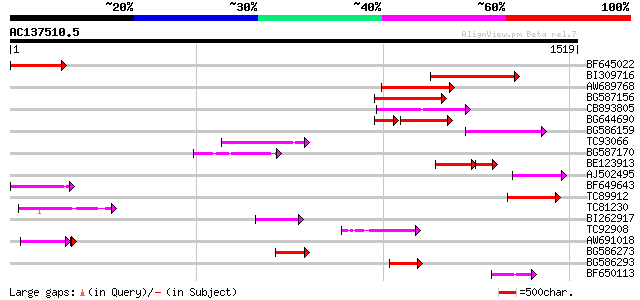
Score E
Sequences producing significant alignments: (bits) Value
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 312 5e-85
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 235 9e-62
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 205 1e-52
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 179 6e-45
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 172 9e-43
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 138 5e-42
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 162 7e-40
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 144 3e-34
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 133 6e-31
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 94 4e-29
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 112 1e-24
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 108 1e-23
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 107 3e-23
TC81230 103 4e-22
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 100 6e-21
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 98 3e-20
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 89 9e-19
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 90 8e-18
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 83 9e-16
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 79 1e-14
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 312 bits (800), Expect = 5e-85
Identities = 151/151 (100%), Positives = 151/151 (100%)
Frame = -2
Query: 1 MGENSGDGGKKVDDNSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSF 60
MGENSGDGGKKVDDNSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSF
Sbjct: 453 MGENSGDGGKKVDDNSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSF 274
Query: 61 QAKRKYGFVEGKIPKPTTPEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTH 120
QAKRKYGFVEGKIPKPTTPEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTH
Sbjct: 273 QAKRKYGFVEGKIPKPTTPEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTH 94
Query: 121 LKQRFCVVNGARICQLKASLGECKQGKGEEV 151
LKQRFCVVNGARICQLKASLGECKQGKGEEV
Sbjct: 93 LKQRFCVVNGARICQLKASLGECKQGKGEEV 1
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 235 bits (600), Expect = 9e-62
Identities = 118/239 (49%), Positives = 167/239 (69%)
Frame = +2
Query: 1128 VCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLI 1187
VC L+KS+YGLKQA R W++KL +L +G++QS SD+SLFT + +LVYVDD++
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 1188 IAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGL 1247
+AGNDI+ ++ K +L F +KDLG L+YFLGLEVAR+ +GI L QRKY LE+++++G
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 1248 LGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQK 1307
L K P + KL D YRRLIG+LIYL+ TRPD++++V LSQF+ K
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 1308 PCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLL 1366
P + H++AA+RV++YLK P +G+ + S LKL + DSDWA+CP TR+S+TG+ V L
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 205 bits (521), Expect = 1e-52
Identities = 97/195 (49%), Positives = 133/195 (67%)
Frame = +1
Query: 997 VKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVV 1056
+ D W AM+ E +AL DN+TW + LPP KKA+G KWVY++K + DGS+ + KARLV
Sbjct: 82 LSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVA 261
Query: 1057 FGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKV 1116
G Q G DY ETF+PV K VT+R L +A KWE+ Q+D++NAFL+G L+EEVYM
Sbjct: 262 KGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQ 441
Query: 1117 PPGFKNTDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQ 1176
P GF+ + +LVC+L KSLYGLKQAPR W+ L +A ++GF +S D SL ++
Sbjct: 442 PQGFEAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGAC 621
Query: 1177 INVLVYVDDLIIAGN 1191
I + +YVDD++I G+
Sbjct: 622 IYLXIYVDDILITGS 666
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 179 bits (455), Expect = 6e-45
Identities = 92/195 (47%), Positives = 128/195 (65%), Gaps = 1/195 (0%)
Frame = -1
Query: 977 HRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWV 1036
H F+ + P +++EA++D W++++ E A+ N+TW +LP GKKA+ S+W+
Sbjct: 597 HCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWI 418
Query: 1037 YKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQ 1096
+ IK+ +DGSIER K RLV G G DY ETFAPVAK+ T+R L++A W + Q
Sbjct: 417 FTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQ 238
Query: 1097 MDVHNAFLHGDLEEEVYMKVPPGFKN-TDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKR 1155
MDV NAFL G+LE+EVYM PPG ++ V RLKK++YGLKQ+PR W+ KL T L
Sbjct: 237 MDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNG 58
Query: 1156 YGFVQSYSDYSLFTL 1170
GF +S D++LFTL
Sbjct: 57 RGFRKSELDHTLFTL 13
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 172 bits (436), Expect = 9e-43
Identities = 100/253 (39%), Positives = 150/253 (58%), Gaps = 3/253 (1%)
Frame = +3
Query: 984 VTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHS 1043
+T +P F+EAVK WR +M NE++A E N TW + L G K +G KW++K K +
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 1044 DGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAA-IKKWEVHQMDVHNA 1102
+G IE+ KARLV G+ Q G+DY E FAPVA+ T+R +A+AA IK+ V + A
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KA 383
Query: 1103 FLHGDLEEEVYMKVPPGFKNTDPNLVC-RLKKSLYGLKQAPRCWFAKLVTALKRYGFVQS 1161
H +E + + + ++ R+K++LYGLKQAPR W++++ + GF +
Sbjct: 384 --HSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKC 557
Query: 1162 YSDYSLFT-LHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLG 1220
+++LF L G + + +YVDDLI GND + FK + F+M DLG + YFLG
Sbjct: 558 PYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLG 737
Query: 1221 LEVARNHEGIYLC 1233
+EV +N +GIY+C
Sbjct: 738 VEVTQNEKGIYIC 776
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 138 bits (347), Expect(2) = 5e-42
Identities = 71/141 (50%), Positives = 101/141 (71%), Gaps = 1/141 (0%)
Frame = -2
Query: 1047 IERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHG 1106
I R K++LVV G++Q EGIDYDE F+PVA+M +R +A AA ++++QMDV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1107 DLEEEVYMKVPPGFKNTD-PNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDY 1165
DL+EEV++K PPGF++ + PN V RL K+LYGLKQAPR W+ +L L + GF + D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1166 SLFTLHRGEIQINVLVYVDDL 1186
+LF L R + + VYVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 53.1 bits (126), Expect(2) = 5e-42
Identities = 23/65 (35%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Frame = -3
Query: 978 RNFVA--AVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKW 1035
RN V+ A + EP N KEA++D+ W ++M+ E+ E ++ W + P GK +G++W
Sbjct: 627 RNLVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRW 448
Query: 1036 VYKIK 1040
V++ K
Sbjct: 447 VFRNK 433
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 162 bits (411), Expect = 7e-40
Identities = 80/217 (36%), Positives = 130/217 (59%)
Frame = +1
Query: 1221 LEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRL 1280
+EV +N EGIY+CQRKY ++++ G+ + + P+ KL D Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1281 IGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELK 1340
+G L+YL+ TRPDL Y + ++S+FM P E H A RV+RYL GI+ + + K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1341 LEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCEL 1400
LE + DSD+A R+S +G+V +L VSW +KKQP V+ S+ +AE+ + A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1401 KWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFH 1437
W++++L LG + S + +YCD+ S + +++NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 144 bits (363), Expect = 3e-34
Identities = 86/242 (35%), Positives = 131/242 (53%), Gaps = 6/242 (2%)
Frame = +1
Query: 567 SFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETM 626
SF + + + + +H DLWG + S G +Y +TI++D+ R VWVY L K E
Sbjct: 61 SFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPT 240
Query: 627 FLNFVAFVDRQFDKKIKKVRSDNGTEF--NCLRDYFFNNGIVFETSCVGTPQQNGRVERK 684
F + V+ Q K +KK+ +DN EF + ++ N+GI + PQQNG ER
Sbjct: 241 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERM 420
Query: 685 HQHIMNVARALRFQGHL--PMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFD 742
+ ++ AR + L W E TAC+L+NR+P S L++K P + G + +
Sbjct: 421 IRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYS 600
Query: 743 NMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLY--DLESKEYIVSRDVKF 800
N++IFGC YA DG K A R+ +CIF+ Y KG++L+ D +S++ I+SRDV F
Sbjct: 601 NLRIFGCPAYA--LVNDG-KLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTF 771
Query: 801 YE 802
E
Sbjct: 772 NE 777
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 133 bits (334), Expect = 6e-31
Identities = 84/245 (34%), Positives = 127/245 (51%), Gaps = 8/245 (3%)
Frame = -3
Query: 492 LIGAGERIDGLYFFRGVPKVHALMVEGDSAMDL-----WHKRLGHPSEKVLKF-IPHVSQ 545
LIG G LY + V S+ L WH RLGHP + L +P V
Sbjct: 695 LIGEGVTKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLPGVVF 516
Query: 546 HSRSKNNRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIV 605
N+ C+ C K ++ FP + + F+L++ DLW + + S +Y++T +
Sbjct: 515 E-----NKNCEACILGKHCKNVFPRTSTVYENCFDLIYTDLWTA-PSLSRDNHKYFVTFI 354
Query: 606 NDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFN--CLRDYFFNN 663
++ S+ W+ L+ +K + F NF A+V + KIK +RSDNG E+ + + ++
Sbjct: 353 DEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHH 174
Query: 664 GIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSS 723
GI+ +TSC TPQQNG +RK++H+M VAR+L FQ + V TACYLIN P+
Sbjct: 173 GILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTACYLINWIPTK 21
Query: 724 VLNYK 728
VL K
Sbjct: 20 VLRIK 6
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 93.6 bits (231), Expect(2) = 4e-29
Identities = 46/113 (40%), Positives = 73/113 (63%), Gaps = 1/113 (0%)
Frame = +1
Query: 1140 QAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVL-VYVDDLIIAGNDIAALKI 1198
Q+PR WF + +K++G++Q +D+++F H ++ +L VYVDD+ + G+ +K
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1199 FKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAK 1251
K L F +KDLG LKYFLG+EVAR +G + QRKY L+++ ET ++G K
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCK 339
Score = 54.7 bits (130), Expect(2) = 4e-29
Identities = 27/58 (46%), Positives = 38/58 (64%)
Frame = +2
Query: 1248 LGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFM 1305
L KP++ PM+ KL + L D Y+RL+G+LIYLS TRPD+++ V +SQFM
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 112 bits (280), Expect = 1e-24
Identities = 56/146 (38%), Positives = 87/146 (59%)
Frame = +2
Query: 1347 SDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQL 1406
SDWA TR+S +G+ L +SW +KKQP V+ S+AEAEY + + WL+++
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1407 LGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPT 1466
L + + ++YCD+KSA+ +++NPVFH R+KHI+ H +R+ + + Y PT
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1467 SVQLADIFTKALGKAQFEFLLRKLGI 1492
++ADIFTK L F L + LG+
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 108 bits (271), Expect = 1e-23
Identities = 57/175 (32%), Positives = 91/175 (51%), Gaps = 3/175 (1%)
Frame = +1
Query: 3 ENSGDGGKKVDDNSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQA 62
E S + K +N +NS+ +++ +D+PG+++ P +L G NY WSR++ + A
Sbjct: 55 EKSNNNSNK-SNNKNNSDMDSSNPYYIHPSDHPGHLLVPTKLNGTNYSSWSRSMVHALTA 231
Query: 63 KRKYGFVEGKIPKPT---TPEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWT 119
K K GF+ G I P+ P + W SM+++WL +++EP L + + A +W
Sbjct: 232 KNKVGFINGSIKTPSEVDQPAEYALWNRCNSMILSWLTHSVEPDLAKGVIHAKTACQVWE 411
Query: 120 HLKQRFCVVNGARICQLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTC 174
K +F N I Q++ SL QG S YF ++ +WDEL TY PTC
Sbjct: 412 DFKDQFSQKNIPAIYQIQKSLASLSQGT-VSASTYFTKIKGLWDELETYRTLPTC 573
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 107 bits (268), Expect = 3e-23
Identities = 54/142 (38%), Positives = 91/142 (64%)
Frame = +1
Query: 1335 SDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMA 1394
+ E LEG+ D+D+A TR+SL+G+V L + +SWK +Q V+ S+ +AEY +
Sbjct: 70 AQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKANQQSVVTLSTTQAEYIAFV 249
Query: 1395 MTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAV 1454
+ WLK ++G+LG++ + ++++CDS+SA+H+A + V+HERTKHI+ HF+RD +
Sbjct: 250 EGVKDAIWLKGMIGELGIT-QEYVKIHCDSQSAIHLANHQVYHERTKHIDIRLHFIRDMI 426
Query: 1455 VAGIICPLYVPTSVQLADIFTK 1476
+ I + + AD+FTK
Sbjct: 427 ESKEIVVEKMASEENPADVFTK 492
>TC81230
Length = 958
Score = 103 bits (258), Expect = 4e-22
Identities = 73/275 (26%), Positives = 119/275 (42%), Gaps = 13/275 (4%)
Frame = +1
Query: 25 LVFFLGSNDNPGNVITPIQ--LRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTT---- 78
++FFL S V PI L G NY+ W+ ++ + +R + +V G PT
Sbjct: 175 VLFFLDSLMEKLEVTPPISIILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDD 354
Query: 79 -----PEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARI 133
+KLE+W + +I W NT PS+ +++A+ +W HLKQR+ + + +
Sbjct: 355 TADAFADKLEEWDSKNHQIITWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQ 534
Query: 134 CQLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAE 193
QL L KQ G+ V + ++ IW++L + CE + D T +
Sbjct: 535 YQLLKDLSNLKQQSGQPVYEFLAQMEVIWNQLTS--------CEPSLKDATDMKTYETHR 690
Query: 194 D--YLHHFLMGLDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVM 251
+ L FLM L Y +R++ L Q+PLP ++ A + +E Q
Sbjct: 691 NRVRLIQFLMALTDEYEPVRASSLHQNPLPTLENALPCLKSEETRLQ------------- 831
Query: 252 AFKVAPDTRGKSKLVDNSDKFCTHCNREGHDERTC 286
V P + +N+ K C HC + GH C
Sbjct: 832 --LVPPKADLAFAVTNNATKPCRHCQKSGHSFSDC 930
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 100 bits (248), Expect = 6e-21
Identities = 51/129 (39%), Positives = 75/129 (57%), Gaps = 1/129 (0%)
Frame = +1
Query: 659 YFFNNGIVFETSCVG-TPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLI 717
+F N ++ S V TPQQNG ER ++ ++ RA+ + FW E V TACY+I
Sbjct: 37 HFVNKRVL*GNSXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVI 216
Query: 718 NRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPY 777
NR+PS+V++ KTP E GK + ++ +FGC Y ++ K +SRKCIF+GY
Sbjct: 217 NRSPSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYAD 396
Query: 778 GKKGWKLYD 786
KG+ L+D
Sbjct: 397 NVKGYXLWD 423
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 97.8 bits (242), Expect = 3e-20
Identities = 72/212 (33%), Positives = 109/212 (50%)
Frame = +2
Query: 888 EVDGADVVVMQEDDVEPGEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKS 947
+V+ +D+ +QED GE + E+ E+ +G + K+K+ +H + S
Sbjct: 26 QVEASDLEEIQEDFKSHGE----VQEESNYIEEI-KGFQEPTQLRKIKE*ESHLVTLNSS 190
Query: 948 SKSSSAQEDASGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMR 1007
SKS Y I ++V FS +HR +AA+T+ EP N+ +A + W AM
Sbjct: 191 SKS-----------YHIFHYVGYS-FSAKHRASLAAITSNIEPKNYVQAAQ*QEWLAAME 334
Query: 1008 NEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDY 1067
EIQ LE+N T +E L GKK + + VYKI H ++G IE+ KA+LV Q+EG D+
Sbjct: 335 QEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF 514
Query: 1068 DETFAPVAKMVTVRTFLAVAAIKKWEVHQMDV 1099
+ K R L +AA K ++H MDV
Sbjct: 515 *D-LCLSNKDDNCRCLLTIAAAKG*QLHLMDV 607
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 89.0 bits (219), Expect(2) = 9e-19
Identities = 44/134 (32%), Positives = 76/134 (55%), Gaps = 1/134 (0%)
Frame = +2
Query: 29 LGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPE-KLEDWKA 87
L +D PG + L G NY EWSR++ S AK K G ++G + P+ + KL WK
Sbjct: 164 LHHSDTPGISLVNQPLNGGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADDPKLPLWKR 343
Query: 88 VQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQGK 147
+++ W+L++IEP + ++ + D A ++W+ L RF + +RI Q++ + EC+QG
Sbjct: 344 CNDLVLTWILHSIEPDIARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQG- 520
Query: 148 GEEVSAYFGRLSRI 161
+S Y+ +L +
Sbjct: 521 SLLISDYYTKLKSL 562
Score = 24.3 bits (51), Expect(2) = 9e-19
Identities = 8/16 (50%), Positives = 11/16 (68%)
Frame = +3
Query: 162 WDELVTYVKKPTCKCE 177
WDEL +Y + TC C+
Sbjct: 564 WDELGSYQEPITCSCD 611
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 89.7 bits (221), Expect = 8e-18
Identities = 43/90 (47%), Positives = 58/90 (63%)
Frame = -2
Query: 713 ACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIF 772
ACYLINR P+ VL + P+E L + P M++FGCLCY +K +RSRK +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 773 VGYPYGKKGWKLYDLESKEYIVSRDVKFYE 802
+GY +KG+K YD E++ +VSRDVKF E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 82.8 bits (203), Expect = 9e-16
Identities = 40/90 (44%), Positives = 60/90 (66%)
Frame = +2
Query: 1017 ETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAK 1076
+T + K P G K +G +W+YKIK + DG++ + KARLV G+ + +GID+DE FAPV +
Sbjct: 53 QTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVR 232
Query: 1077 MVTVRTFLAVAAIKKWEVHQMDVHNAFLHG 1106
+ T+ LA+AA +H +DV AFL+G
Sbjct: 233 IETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 79.0 bits (193), Expect = 1e-14
Identities = 47/123 (38%), Positives = 72/123 (58%), Gaps = 3/123 (2%)
Frame = +1
Query: 1291 RPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLR--SDSEL-KLEGWCDS 1347
RPD+ YSV ++S+FM P + H AA R++RY++ G+L + SE+ +L + DS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1348 DWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLL 1407
DW RRS +G+V + + +SW TKKQP + SS EAEY + T + WL ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1408 GDL 1410
+L
Sbjct: 472 KEL 480
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,155,801
Number of Sequences: 36976
Number of extensions: 767272
Number of successful extensions: 4056
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 3857
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3991
length of query: 1519
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1410
effective length of database: 4,984,343
effective search space: 7027923630
effective search space used: 7027923630
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137510.5


