

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137077.8 - phase: 0
(1091 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
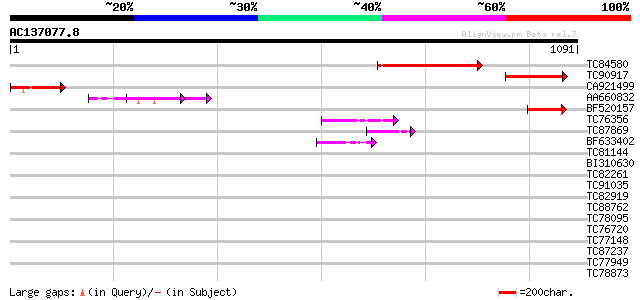
Score E
Sequences producing significant alignments: (bits) Value
TC84580 weakly similar to GP|20466528|gb|AAM20581.1 G2484-1 prot... 258 1e-68
TC90917 similar to GP|20466528|gb|AAM20581.1 G2484-1 protein {Ar... 228 1e-59
CA921499 similar to PIR|E71442|E71 hypothetical protein - Arabid... 133 3e-31
AA660832 GP|1405736|emb trypsin inhibitor cme precursor {Hordeum... 93 5e-19
BF520157 weakly similar to GP|20466528|gb G2484-1 protein {Arabi... 86 1e-16
TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown prot... 57 5e-08
TC87869 similar to GP|23504577|emb|CAD51456. chromosome condensa... 47 5e-05
BF633402 similar to PIR|C86226|C862 protein T31J12.4 [imported] ... 44 4e-04
TC81144 similar to PIR|T04462|T04462 hypothetical protein F4D11.... 40 0.006
BI310630 similar to GP|18087548|gb AT5g55600/MDF20_4 {Arabidopsi... 39 0.011
TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity... 38 0.024
TC91035 similar to PIR|C84650|C84650 hypothetical protein At2g25... 37 0.032
TC82919 weakly similar to GP|8843783|dbj|BAA97331.1 gb|AAC80581.... 37 0.054
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 37 0.054
TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein... 36 0.093
TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {... 35 0.21
TC77148 ENOD20 35 0.21
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 35 0.21
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 34 0.27
TC78873 similar to GP|21740530|emb|CAD41509. OSJNBa0029H02.25 {O... 34 0.35
>TC84580 weakly similar to GP|20466528|gb|AAM20581.1 G2484-1 protein
{Arabidopsis thaliana}, partial (7%)
Length = 679
Score = 258 bits (658), Expect = 1e-68
Identities = 133/204 (65%), Positives = 159/204 (77%), Gaps = 2/204 (0%)
Frame = +2
Query: 709 KNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEK 768
KNKK +T TV+ PASGETSV AW LRPSLIW DG+W++ KVGAN+S T++GDTPHEK
Sbjct: 2 KNKKAKT-FTVYFPASGETSVFDAWLLRPSLIWNDGKWIESPKVGANNSPTNEGDTPHEK 178
Query: 769 RPKLGSNAVEV-KGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQ 827
RPKLG+ A E+ KGKDK SK DAA+SANP E+R +NLTE++ VFN+GKS+ NE K D
Sbjct: 179 RPKLGNPAQELAKGKDKASKGTDAAKSANPTELRLVNLTEDDKVFNVGKSNKNEKKPDVH 358
Query: 828 RQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVND-KNDSVKIANFSMPQG 886
R RSGLQKEG +VIFGVPKPGK KFMEVSKHYVA G+S++ND N SVK AN S+P
Sbjct: 359 RLARSGLQKEGPRVIFGVPKPGKNTKFMEVSKHYVADGTSRINDGGNGSVKFANSSIPNA 538
Query: 887 SELRGWRNSSKNDSKEKLGADSKP 910
S R W++SS +D+KEK AD KP
Sbjct: 539 SXSRSWKDSSIHDAKEKPRADFKP 610
>TC90917 similar to GP|20466528|gb|AAM20581.1 G2484-1 protein {Arabidopsis
thaliana}, partial (3%)
Length = 579
Score = 228 bits (581), Expect = 1e-59
Identities = 116/119 (97%), Positives = 117/119 (97%)
Frame = +3
Query: 954 RVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKA 1013
RVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKA
Sbjct: 3 RVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKA 182
Query: 1014 LNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRNIPKGEHPQ 1072
LNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRNIPKG + Q
Sbjct: 183 LNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRNIPKGNNHQ 359
>CA921499 similar to PIR|E71442|E71 hypothetical protein - Arabidopsis
thaliana, partial (3%)
Length = 837
Score = 133 bits (335), Expect = 3e-31
Identities = 81/112 (72%), Positives = 87/112 (77%), Gaps = 5/112 (4%)
Frame = -3
Query: 1 MISAYGGTGTCGRMSGVSAWKGSVV-----KNLILIPQKHLCSQDLAARTSDSTVKQSVL 55
MISAYGGT GR + W+ + K+ P+ L S+ AARTSDSTVKQSVL
Sbjct: 658 MISAYGGTDG-GRNLWENVWRVCMERQRSQKSHPNTPETPLQSRS-AARTSDSTVKQSVL 485
Query: 56 QGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSV 107
QGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSV
Sbjct: 484 QGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSV 329
>AA660832 GP|1405736|emb trypsin inhibitor cme precursor {Hordeum vulgare},
partial (9%)
Length = 651
Score = 93.2 bits (230), Expect = 5e-19
Identities = 73/195 (37%), Positives = 103/195 (52%), Gaps = 10/195 (5%)
Frame = +3
Query: 153 PDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
P ++H+S + ++ T + ++P ++ K V P F+ + NN
Sbjct: 93 PSVSSHISTAVATATSAV-----NVPVTTVEKSVEP-----------LFLPSTPLFSTNN 224
Query: 213 VTVPPAQQSSGPKAKKRKKDVL-SEDHGQKL--LQS----LTPAVASRASTSVSAATPVG 265
V AQ SS PK+KKRKK SED GQK LQS TP ++S ST+ + TPV
Sbjct: 225 AMVSRAQHSSDPKSKKRKKVTTESEDLGQKAIHLQSHLVLSTPVISSHISTAFATETPVL 404
Query: 266 NVPMSSVEK-SVVSVSPLA--DQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAA 322
NV + K S V +S + K K+++SDESL K++EAR++A E SALSAAA
Sbjct: 405 NVASNCCGKVSSVCISSIFC*SPQKWLGMFXKKVMSDESLTKIEEARINAXETSALSAAA 584
Query: 323 VNHSLELWNQLDKHK 337
VN + +W QLD K
Sbjct: 585 VNXXMXIWKQLDNQK 629
Score = 48.1 bits (113), Expect = 2e-05
Identities = 49/175 (28%), Positives = 80/175 (45%), Gaps = 11/175 (6%)
Frame = +3
Query: 225 KAKKRKKDVL-SEDHGQKL--LQS----LTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
K+KKRKK SED GQK LQS TP+V+S ST+V+ AT NVP+++VEKSV
Sbjct: 3 KSKKRKKVTTESEDLGQKAMHLQSHLVLSTPSVSSHISTAVATATSAVNVPVTTVEKSVE 182
Query: 277 ---VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQL 333
+ +PL N + SD K K+ +E+ + +H + +
Sbjct: 183 PLFLPSTPLF-STNNAMVSRAQHSSDPKSKKRKKVTTESEDLGQKAIHLQSHLVLSTPVI 359
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEAL 388
H ++ F ++ ++ ++ + + F K+M+DE+L
Sbjct: 360 SSHISTAFATETPVLNVASNCCGKVSSVCISSIFC*SPQKWLGMFXKKVMSDESL 524
>BF520157 weakly similar to GP|20466528|gb G2484-1 protein {Arabidopsis
thaliana}, partial (3%)
Length = 525
Score = 85.5 bits (210), Expect = 1e-16
Identities = 48/82 (58%), Positives = 57/82 (68%), Gaps = 7/82 (8%)
Frame = +1
Query: 996 GKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSK 1055
GK APA +L KG KALN P STSE + LEPRRSNR+IQPTSRLLEG+QSSL+++K
Sbjct: 1 GKPAPAGGRLGKGEVEKALNGNPLKSTSE-EVLEPRRSNRKIQPTSRLLEGIQSSLIITK 177
Query: 1056 IPSVS-------HNRNIPKGEH 1070
PS S HNRN +G +
Sbjct: 178 TPSASHEKSQKNHNRNTSRGNN 243
>TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown protein
{Arabidopsis thaliana}, partial (36%)
Length = 2619
Score = 56.6 bits (135), Expect = 5e-08
Identities = 43/153 (28%), Positives = 64/153 (41%), Gaps = 6/153 (3%)
Frame = +3
Query: 601 GSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGP---LKEWVSLE---CEG 654
GS VEV + G + WF +++ KV V Y + E L+EW+
Sbjct: 1182 GSNVEVLSQDSGIRGCWFRASVIKRHKDKVKVQYHDIQDAEDEANNLEEWILASRPVVPD 1361
Query: 655 DKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDE 714
D R+ + L KR + +GD VG VDAW + W EG++ +K D+
Sbjct: 1362 DLGLRVEERTKIRPL----LEKRGISFVGD----VGYIVDAWWHDGWWEGIVVQKESDDK 1517
Query: 715 TTLTVHIPASGETSVLRAWNLRPSLIWKDGQWL 747
V+ P + S+ NLR S W W+
Sbjct: 1518 --YHVYFPGEKKMSIFGPCNLRHSRDWTGNGWV 1610
>TC87869 similar to GP|23504577|emb|CAD51456. chromosome condensation
protein putative {Plasmodium falciparum 3D7}, partial
(0%)
Length = 1154
Score = 46.6 bits (109), Expect = 5e-05
Identities = 27/95 (28%), Positives = 44/95 (45%)
Frame = +1
Query: 687 WSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
+ +G VDAW + W E +IT + + T V+ P + ++ +LR S W D W
Sbjct: 34 FEIGAAVDAWCGDGWWESIITAVDASENGTCQVYSPGEEKFLLVEKRDLRISQDWIDNMW 213
Query: 747 LDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKG 781
++ +G D + P RPK +N+ V G
Sbjct: 214 VNI--LGKPDICSFLSSNP--PRPKFQANSAVVNG 306
>BF633402 similar to PIR|C86226|C862 protein T31J12.4 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 427
Score = 43.5 bits (101), Expect = 4e-04
Identities = 34/118 (28%), Positives = 52/118 (43%), Gaps = 1/118 (0%)
Frame = +1
Query: 590 SENLEENTFKEGSLVEVFKDEEGHKAAWFMGNIL-SLKDGKVYVCYTSLVAVEGPLKEWV 648
S + FK G+LVE+ D++G + +WF G I+ L + K V Y +L+ E K
Sbjct: 130 SSSAASEFFKPGTLVEISSDDDGFRGSWFTGKIVRRLANNKFMVEYDNLMEDEAGTKHL- 306
Query: 649 SLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVI 706
+ ++ RP+ L E R + GD VDA+ + W EG I
Sbjct: 307 ------KESLKLHQLRPV--LPTEANR----------VFKFGDEVDAYHNDGWWEGHI 426
>TC81144 similar to PIR|T04462|T04462 hypothetical protein F4D11.160 -
Arabidopsis thaliana, partial (13%)
Length = 1102
Score = 39.7 bits (91), Expect = 0.006
Identities = 38/113 (33%), Positives = 49/113 (42%)
Frame = +1
Query: 84 SPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRG 143
SP S PT S +A G V PL P P P+NF SQ+P
Sbjct: 130 SPFGSRPTGPPGSF--AAPVSGVGVPPPGGSPPLRP-SGPPPQNFGARP----SQSPFNA 288
Query: 144 PWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSG 196
P SP AP N + +A PSS + ++PPS+ + + GPPA S G
Sbjct: 289 PPSQSPFNAPPNQSPFNAPPSSAPPGMPPT--NVPPSNLLSN---GPPAFSGG 432
>BI310630 similar to GP|18087548|gb AT5g55600/MDF20_4 {Arabidopsis thaliana},
partial (19%)
Length = 730
Score = 38.9 bits (89), Expect = 0.011
Identities = 40/187 (21%), Positives = 67/187 (35%), Gaps = 6/187 (3%)
Frame = +1
Query: 524 VRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKPVDMVLGSESETQDPS 583
V D + ++R + + +I G Q V + +P +L + P
Sbjct: 226 VGDDAKRGAKRSRSVTNGREGVRKLIRSNPMMGYQTFQVVNYARPDRRLLSLKKVDCKPW 405
Query: 584 FTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGP 643
F T+K +EV + G + WF I+ + ++ V Y + +G
Sbjct: 406 FNP----------TYKVDDKIEVLSQDSGIRGCWFRCTIVQVARKQLKVQYDDVQDEDGS 555
Query: 644 --LKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMG----DYAWSVGDRVDAWI 697
L+EW+ P + A+P + R R A + VG VDAW
Sbjct: 556 GNLEEWI---------PAFKLAKPDKLEMRQPGRSTIRPAPPLEEQELIVEVGTAVDAWW 708
Query: 698 QESWREG 704
+ W EG
Sbjct: 709 SDGWGEG 729
>TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity to
phytocyanin/early nodulin-like protein~gene_id:K19P17.3,
partial (7%)
Length = 740
Score = 37.7 bits (86), Expect = 0.024
Identities = 33/111 (29%), Positives = 50/111 (44%), Gaps = 3/111 (2%)
Frame = +1
Query: 68 SSKATPTIANPL-IPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPR 126
SS TP+ +P P +SP + LP+L S L+P P SPSP
Sbjct: 406 SSSPTPSPVSPSESPSNSPSFFLPSLLT----------------SPELSP-SPSSSPSPS 534
Query: 127 NFLGHSTSWISQAPLRGPWIGSPTPAPDNN--THLSASPSSDTIKLASVKG 175
+ S + +S P WI +P+PAP +N S++P ++ + S G
Sbjct: 535 PVISPSPTVLSTPP-HSSWISAPSPAPSSNQMNSQSSNPKHHSVVIWSTVG 684
>TC91035 similar to PIR|C84650|C84650 hypothetical protein At2g25590
[imported] - Arabidopsis thaliana, partial (7%)
Length = 709
Score = 37.4 bits (85), Expect = 0.032
Identities = 32/150 (21%), Positives = 51/150 (33%), Gaps = 2/150 (1%)
Frame = +2
Query: 600 EGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPR 659
+GS VE+ + +GH W I+S V Y + PL V + PP
Sbjct: 188 KGSKVEILVNTQGHGVEWHCARIISGNGHSYNVEYDNPSVTAKPLSNRVPRKFIRPCPPA 367
Query: 660 IRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTV 719
I +W D V+ W W+E + ++ + V
Sbjct: 368 IENIG---------------------SWECNDTVEVWDAGCWKEATVLTDMTEEFYLVRV 484
Query: 720 HIPASGETSVLRAWNL--RPSLIWKDGQWL 747
H G L+ + R W++GQW+
Sbjct: 485 H----GSCMELKVHKILTRICQSWQNGQWI 562
>TC82919 weakly similar to GP|8843783|dbj|BAA97331.1
gb|AAC80581.1~gene_id:MZN1.7~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 1227
Score = 36.6 bits (83), Expect = 0.054
Identities = 45/172 (26%), Positives = 68/172 (39%), Gaps = 17/172 (9%)
Frame = +2
Query: 595 ENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVE--GPLKEWVSLE- 651
++ K VEV EEG +W G ++ + K +V Y +++ G E VS+
Sbjct: 35 QSKLKINERVEVKSFEEGFLGSWHPGTVIDSEKLKRHVQYENILNDNGLGNFVEIVSVSS 214
Query: 652 -CEGD-----KPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGV 705
+GD K RIR PL + + G VD QE+W EGV
Sbjct: 215 VLDGDIGSLSKRGRIRPVPPLVEFEKCDLK-------------YGLCVDVNYQEAWWEGV 355
Query: 706 ITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKD--------GQWLDF 749
+ +K E TV P G+ + +R + W + G+WL F
Sbjct: 356 VNDKCDGMEER-TVFFPDLGDEMKVGIQEMRITQDWDEVTEKWEPRGKWLLF 508
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 36.6 bits (83), Expect = 0.054
Identities = 39/131 (29%), Positives = 54/131 (40%), Gaps = 3/131 (2%)
Frame = +3
Query: 62 SPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQ 121
+P G + + NP P SSP S P + S +S A S V + + P
Sbjct: 36 TPSGNGIAPSPYQFNNPTSPSSSPSASSPVSTTTSPPASTPA-SSPVSTTTSPPASTPAS 212
Query: 122 SPSPRNFLGHSTSWISQAPLRGPW-IGSPTPAPDNNTHLSA--SPSSDTIKLASVKGSLP 178
SP P +TS + P P SPT +P + +A SPSS T S + P
Sbjct: 213 SPVPT-----TTSPPAPTPASSPVSTNSPTASPAGSLPAAATPSPSSTTAGSPSPSSTPP 377
Query: 179 PSSSIKDVTPG 189
+ S + TPG
Sbjct: 378 GTRSSNETTPG 410
>TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein At2g36090
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1467
Score = 35.8 bits (81), Expect = 0.093
Identities = 30/112 (26%), Positives = 43/112 (37%), Gaps = 4/112 (3%)
Frame = +3
Query: 117 LHPYQSPS----PRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLAS 172
L P+Q + P N H WI Q+P P +PT +PD H S
Sbjct: 189 LKPFQYSNTNYQPNNI--HIHLWIHQSPPSTPTYSNPTSSPDLTAHHS------------ 326
Query: 173 VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGP 224
PPS P PP ++ Q+T G S ++++ P + S P
Sbjct: 327 -----PPS------PPPPPTFTTSAQNTTYGKKSSPPHGHLSITPPPRHSSP 449
>TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (51%)
Length = 1290
Score = 34.7 bits (78), Expect = 0.21
Identities = 41/158 (25%), Positives = 64/158 (39%)
Frame = +1
Query: 69 SKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNF 128
S T I+N ++ ++ P +SL L L + A + S + L+ SPSP
Sbjct: 448 SDPTIQISNSILSMAIPRFSLVFLLLSFLVNIASSADSPAPTPATNSSLN---SPSP--- 609
Query: 129 LGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTP 188
T + +P P +PTP P + H + P+ S S PSSS +P
Sbjct: 610 ----TPIPTPSPANSP--PAPTPTPTPSPHSDSPPAPSPDNSPSSSPSPSPSSS-PAPSP 768
Query: 189 GPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKA 226
A ++ + T +G D + + SSG KA
Sbjct: 769 DEAADNNAISHTGIGEDGKSSGGGM-------SSGKKA 861
>TC77148 ENOD20
Length = 1108
Score = 34.7 bits (78), Expect = 0.21
Identities = 28/77 (36%), Positives = 34/77 (43%)
Frame = +3
Query: 119 PYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLP 178
P SPSP ++ I R P SP+P+ + S SPS SV SL
Sbjct: 519 PSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSV-ASLA 695
Query: 179 PSSSIKDVTPGPPASSS 195
PSSS D +P P S S
Sbjct: 696 PSSSPSDESPSPAPSPS 746
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 34.7 bits (78), Expect = 0.21
Identities = 51/289 (17%), Positives = 107/289 (36%), Gaps = 6/289 (2%)
Frame = +1
Query: 742 KDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVK---GKDKMSKNIDAAESANPD 798
KDG+ + D + + K + E+K G +K + I +S +
Sbjct: 181 KDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQEN 360
Query: 799 EMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQ---KEGSKVIFGVPKPGKKRKFM 855
E S ++++ +N++ D + QV + K+G++ G + G+K +
Sbjct: 361 EETSETNSKDK-----ENEESNQNGSDAKEQVGENHEQDSKQGTEETNGT-EGGEKEEHD 522
Query: 856 EVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFG 915
++ + + + +KN+ + N+S S S ++ +K + D K K +F
Sbjct: 523 KIKEDTSSDNQVQDGEKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFDKKEKNEFE 702
Query: 916 KPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSS 975
N ++ S N + K+ +Q+E+ + S +D Q ++
Sbjct: 703 LES--QKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQEQNNT 876
Query: 976 QATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE 1024
T T + ++ + + D K ALN P S+
Sbjct: 877 TKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDSK 1023
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 34.3 bits (77), Expect = 0.27
Identities = 36/136 (26%), Positives = 51/136 (37%), Gaps = 2/136 (1%)
Frame = +3
Query: 70 KATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRN-- 127
K T + +P P SSP S LS S S S + + P SP P +
Sbjct: 465 KLTLVVISPRTPKSSPSPSAGGLSPPS-PSPTTTTPSPSGSPPSPVAIPPASSPVPTSGP 641
Query: 128 FLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVT 187
+ +S P GP SP+P +T + P + + A +L P+
Sbjct: 642 TASSPSPVVSTPPAGGPMASSPSPVV--STPPAGGPMASSPSPAGGPPALSPAGGPSTAA 815
Query: 188 PGPPASSSGLQSTFVG 203
GPPA G +T G
Sbjct: 816 GGPPAPGPGGAATSPG 863
>TC78873 similar to GP|21740530|emb|CAD41509. OSJNBa0029H02.25 {Oryza
sativa}, partial (7%)
Length = 623
Score = 33.9 bits (76), Expect = 0.35
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Frame = +3
Query: 148 SPTPAPDNNTHLSASPSSDTIKL-ASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDS 206
SP+ +P + +S S S A VK S PS S + P PP +SSG +
Sbjct: 180 SPSSSPSKSPAISPSAHSPAASPPAPVKNSPSPSPSAINSPPSPPPASSGSPA----AAP 347
Query: 207 QLDANNVTVPPAQQSSGPKAKKR 229
+ ++++ PPA+ S A R
Sbjct: 348 AVTPSSISTPPAEAPSNGAALNR 416
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,176,639
Number of Sequences: 36976
Number of extensions: 405069
Number of successful extensions: 2348
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 2103
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2253
length of query: 1091
length of database: 9,014,727
effective HSP length: 106
effective length of query: 985
effective length of database: 5,095,271
effective search space: 5018841935
effective search space used: 5018841935
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137077.8


