

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
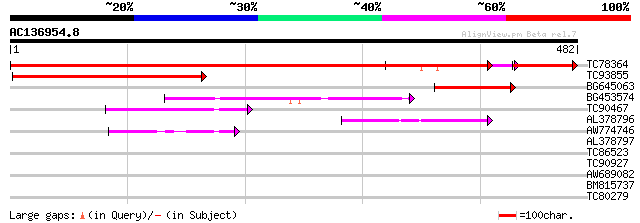
Query= AC136954.8 + phase: 0
(482 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78364 similar to GP|9759266|dbj|BAB09587.1 diacylglycerol kina... 771 0.0
TC93855 similar to GP|13430524|gb|AAK25884.1 unknown protein {Ar... 219 1e-57
BG645063 similar to GP|9759266|dbj| diacylglycerol kinase-like p... 144 6e-35
BG453574 similar to GP|13430776|gb putative diacylglycerol kinas... 127 7e-30
TC90467 similar to GP|10798890|gb|AAG23128.1 calmodulin-binding ... 99 5e-21
AL378796 similar to GP|10798892|gb diacylglycerol kinase {Lycope... 92 6e-19
AW774746 similar to GP|10177058|db diacylglycerol kinase {Arabid... 60 2e-09
AL378797 similar to GP|10798892|gb| diacylglycerol kinase {Lycop... 33 0.32
TC86523 similar to GP|22136030|gb|AAM91597.1 putative protein {A... 31 1.2
TC90927 similar to GP|9279712|dbj|BAB01269.1 cell division prote... 29 3.5
AW689082 29 3.5
BM815737 weakly similar to GP|10177406|dbj cytochrome P-450-like... 28 6.0
TC80279 weakly similar to PIR|F96505|F96505 probable nucellin [i... 28 7.8
>TC78364 similar to GP|9759266|dbj|BAB09587.1 diacylglycerol kinase-like
protein {Arabidopsis thaliana}, partial (76%)
Length = 1889
Score = 771 bits (1991), Expect(2) = 0.0
Identities = 391/411 (95%), Positives = 393/411 (95%), Gaps = 1/411 (0%)
Frame = +3
Query: 1 MESSPSSTTEDSKKIQVRSSLVESIRGCGLSGMRIDKEDLKKQLTLPQYLRFAMRDSIRL 60
MESSPSSTTEDSKKIQVRSSLVESIRGCGLSGMRIDKEDLKKQLTLPQYLRFAMRDSIRL
Sbjct: 180 MESSPSSTTEDSKKIQVRSSLVESIRGCGLSGMRIDKEDLKKQLTLPQYLRFAMRDSIRL 359
Query: 61 QDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLAD 120
QDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLAD
Sbjct: 360 QDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLAD 539
Query: 121 VKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREP 180
VKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREP
Sbjct: 540 VKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREP 719
Query: 181 VPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPES 240
VPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPES
Sbjct: 720 VPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPES 899
Query: 241 TTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDE 300
TTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDE
Sbjct: 900 TTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDE 1079
Query: 301 KPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNI-LRMHIKRVSSSEWEQVAI 359
KPYLASGPIANKIIYSGYS G F + G I +RMHIKRVSSSEWEQVAI
Sbjct: 1080KPYLASGPIANKIIYSGYSVYSGLVFHTLYKLIQV*GD*EIFVRMHIKRVSSSEWEQVAI 1259
Query: 360 PKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLK 410
PKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLK
Sbjct: 1260PKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLK 1412
Score = 112 bits (280), Expect(2) = 0.0
Identities = 53/55 (96%), Positives = 54/55 (97%)
Frame = +2
Query: 428 IAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDGS 482
+ QAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDGS
Sbjct: 1466 LLQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDGS 1630
Score = 61.2 bits (147), Expect = 8e-10
Identities = 43/121 (35%), Positives = 63/121 (51%), Gaps = 7/121 (5%)
Frame = +1
Query: 320 CTQGWFFTPCTSDPGLRGLRNILRMHIKR----VSSSEWEQVAIPKS--VRAIVALNLH- 372
CTQGWFFTPCTS +G + L I + S S+ + + + + +A+ +
Sbjct: 1138 CTQGWFFTPCTS*SRSKGTKKYLCGCISKGLVAQSGSKLQFLKV*GQ*LL*IFIAMEVEE 1317
Query: 373 SYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAA 432
++G +NP +K ++ G + KQGWHASFVMVDLITAKHIA +
Sbjct: 1318 THGVNQNP------NI*RRKALLKLMSPMGAWKYLV*KQGWHASFVMVDLITAKHIASGS 1479
Query: 433 A 433
+
Sbjct: 1480 S 1482
>TC93855 similar to GP|13430524|gb|AAK25884.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 599
Score = 219 bits (559), Expect = 1e-57
Identities = 111/167 (66%), Positives = 135/167 (80%), Gaps = 2/167 (1%)
Frame = +2
Query: 3 SSPSSTTEDSKKIQV-RSSLVESIRGCGLSGMRIDKEDLKKQLTLPQYLRFAMRDSIRLQ 61
SS ++TT D+ K+ RSS+V+SIRGCG+SG RIDKE+LK+ LT+PQYLR +MRDSIRL+
Sbjct: 98 SSSTTTTGDATKLAAARSSIVDSIRGCGISGQRIDKEELKRNLTMPQYLRISMRDSIRLK 277
Query: 62 DPSAGETLYRN-RAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLAD 120
DP+AGE + RAE + AP P+VVFIN RSGGRHGP LKERLQ LMSEEQVFD+ D
Sbjct: 278 DPTAGEFGFGFIRAEESAAVAPLKPIVVFINPRSGGRHGPVLKERLQDLMSEEQVFDVID 457
Query: 121 VKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVL 167
V P EF+ YGL CLE LA GD+CAKETRE++R+MVAGGDG++G VL
Sbjct: 458 VNPREFLQYGLGCLEALAASGDTCAKETRERIRIMVAGGDGSIGLVL 598
>BG645063 similar to GP|9759266|dbj| diacylglycerol kinase-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 762
Score = 144 bits (364), Expect = 6e-35
Identities = 69/69 (100%), Positives = 69/69 (100%)
Frame = +2
Query: 362 SVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVD 421
SVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVD
Sbjct: 14 SVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVD 193
Query: 422 LITAKHIAQ 430
LITAKHIAQ
Sbjct: 194 LITAKHIAQ 220
>BG453574 similar to GP|13430776|gb putative diacylglycerol kinase
{Arabidopsis thaliana}, partial (40%)
Length = 661
Score = 127 bits (320), Expect = 7e-30
Identities = 79/221 (35%), Positives = 110/221 (49%), Gaps = 8/221 (3%)
Frame = +1
Query: 132 ACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGT 191
A LE L GD A T E+LR++VAGGDGT GW+LG + +L+ PP+ VPLGT
Sbjct: 28 ANLENLKVQGDRLAISTMERLRLIVAGGDGTAGWLLGVVCDLKL---SHSPPIATVPLGT 198
Query: 192 GNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISM--PESTTVKP---- 245
GN+L +F WG P + ++ L + ++D+W L + M P+ T P
Sbjct: 199 GNNLPFAFGWGKKNPGTDEQSVLSFLNQVMKAKEMKIDNWHLLMRMKAPKHGTCDPIAPL 378
Query: 246 --PYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPY 303
P+ L + + IEG ++ G ++NYFS+GMDAQV+Y FH R P
Sbjct: 379 ELPHSLHAFHRVSETDELNIEG-----CHTFRGGFWNYFSMGMDAQVSYAFHSERKLHPE 543
Query: 304 LASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRM 344
+ N+ Y+ CTQGWF P RNI M
Sbjct: 544 KFKNQLVNQSTYAKLGCTQGWFMASLFHPPS----RNIAHM 654
>TC90467 similar to GP|10798890|gb|AAG23128.1 calmodulin-binding
diacylglycerol kinase {Lycopersicon esculentum}, partial
(27%)
Length = 700
Score = 98.6 bits (244), Expect = 5e-21
Identities = 53/125 (42%), Positives = 74/125 (58%)
Frame = +3
Query: 82 PTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
P P+VVFIN +SGG+ G L L+++ QV+DL P + + A LE L G
Sbjct: 279 PACPVVVFINTKSGGQLGGELLVTYSSLLNQNQVYDLGVHAPDKVLHQLYANLEKLKHNG 458
Query: 142 DSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNW 201
D A E + +LR++VAGGDGT W+LG +++L+ PP+ VPLGTGN+L +F W
Sbjct: 459 DHYAAEIQNRLRIIVAGGDGTASWLLGVVSDLK---LPHPPPIATVPLGTGNNLPFAFGW 629
Query: 202 GGSFP 206
G P
Sbjct: 630 GKKNP 644
>AL378796 similar to GP|10798892|gb diacylglycerol kinase {Lycopersicon
esculentum}, partial (25%)
Length = 383
Score = 91.7 bits (226), Expect = 6e-19
Identities = 47/128 (36%), Positives = 67/128 (51%)
Frame = +2
Query: 283 SIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNIL 342
S+GMDAQV+Y FH R P + N+ Y+ CTQGWFF P R + +
Sbjct: 2 SMGMDAQVSYAFHSERKMNPEKFKNQLVNQSTYAKLGCTQGWFFAGLMH-PSSRNMAQLT 178
Query: 343 RMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADG 402
++ I + +W+ + IP S+R+IV LNL S+ G NPWG P + + V DG
Sbjct: 179 KVKIMQ-KPGQWQDLNIPPSIRSIVCLNLPSFSGGFNPWGTPNRKKQRDRDLTPPYVDDG 355
Query: 403 RLEIFGLK 410
LE+ G +
Sbjct: 356 LLEVVGFR 379
>AW774746 similar to GP|10177058|db diacylglycerol kinase {Arabidopsis
thaliana}, partial (14%)
Length = 395
Score = 59.7 bits (143), Expect = 2e-09
Identities = 42/111 (37%), Positives = 57/111 (50%)
Frame = +2
Query: 85 PMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSC 144
P+ VFIN R+GG+ GP+L +L L++ VF L+ + E LE+ +
Sbjct: 113 PL*VFINTRNGGQLGPSLYRKLNMLLNTVHVFVLSASQGPE------VGLELFKNV---- 262
Query: 145 AKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDL 195
RV+V GGDGTV WVL + + E PPV I+ L TGNDL
Sbjct: 263 -----PYFRVLVCGGDGTVSWVLDAI*KHT---FESPPPVAIISLCTGNDL 391
>AL378797 similar to GP|10798892|gb| diacylglycerol kinase {Lycopersicon
esculentum}, partial (17%)
Length = 439
Score = 32.7 bits (73), Expect = 0.32
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +1
Query: 445 NAYLQMDGEPWKQPLSKDFSTFV 467
+ ++++DGEPWKQPL D T V
Sbjct: 1 HTFMRIDGEPWKQPLPADDDTVV 69
>TC86523 similar to GP|22136030|gb|AAM91597.1 putative protein {Arabidopsis
thaliana}, partial (82%)
Length = 1988
Score = 30.8 bits (68), Expect = 1.2
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 7/76 (9%)
Frame = +2
Query: 154 VMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAI 213
++ GDG + V+ L + P+GIVP GTGN +++S P A +A+
Sbjct: 734 IVCISGDGILVEVVNGLLQREDWDTAIKTPLGIVPAGTGNGMAKSLLDSVGDPCAIANAV 913
Query: 214 -------KRTLQKASV 222
KR L A++
Sbjct: 914 LAIIRGHKRQLDVATI 961
>TC90927 similar to GP|9279712|dbj|BAB01269.1 cell division protein
FtsH-like {Arabidopsis thaliana}, partial (28%)
Length = 984
Score = 29.3 bits (64), Expect = 3.5
Identities = 12/28 (42%), Positives = 19/28 (67%)
Frame = +2
Query: 347 KRVSSSEWEQVAIPKSVRAIVALNLHSY 374
K S +WEQVAI ++ A+ A+NL ++
Sbjct: 230 KERSKEKWEQVAINEAAMAVAAMNLPNF 313
>AW689082
Length = 590
Score = 29.3 bits (64), Expect = 3.5
Identities = 23/106 (21%), Positives = 44/106 (40%), Gaps = 5/106 (4%)
Frame = -2
Query: 352 SEWEQVAIPKSVRAIVALNLHSYGSGR---NPWGKPKPEYLEKKGFVEADVADGRLEIFG 408
S+W ++ + +R ++++NL + NPW P P YL+ G + + +
Sbjct: 331 SKWRKLPLA-GIRRLMSINLSGHFGNTHDVNPWSTPLPHYLD**GLTDGSCRELA*SMLK 155
Query: 409 LKQGWHASFVMVDLITAKHIAQAAAIRL--ELRGGGWKNAYLQMDG 452
+F ++L+T + I+ L W+N LQ G
Sbjct: 154 CLDVIVCAFR*MELLTEYAVPLKLRIQSTPNLNSSIWRNYILQTQG 17
>BM815737 weakly similar to GP|10177406|dbj cytochrome P-450-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 765
Score = 28.5 bits (62), Expect = 6.0
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = +3
Query: 104 ERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLG 141
+R LMS F L D++PH F +GL C + +G
Sbjct: 582 QRRNWLMSARH-FSLVDMRPHHFQSHGLCCF*LCMKIG 692
>TC80279 weakly similar to PIR|F96505|F96505 probable nucellin [imported] -
Arabidopsis thaliana, partial (42%)
Length = 835
Score = 28.1 bits (61), Expect = 7.8
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 3/74 (4%)
Frame = +1
Query: 46 LPQYLRFAMRDSIRLQDP---SAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPAL 102
LPQ MR S++++DP S ET + + + A P+ V + GRH L
Sbjct: 493 LPQMNSVTMRLSMQIRDPLWGSLSETTFLFNSLMDLLYAQE*PLDVDMIKSILGRHPHLL 672
Query: 103 KERLQQLMSEEQVF 116
++ L EEQVF
Sbjct: 673 QQESLALAMEEQVF 714
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,381,743
Number of Sequences: 36976
Number of extensions: 196675
Number of successful extensions: 959
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 943
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 951
length of query: 482
length of database: 9,014,727
effective HSP length: 100
effective length of query: 382
effective length of database: 5,317,127
effective search space: 2031142514
effective search space used: 2031142514
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136954.8


