

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.7 - phase: 0
(396 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
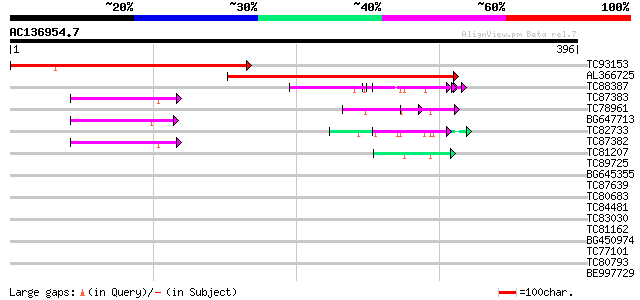
Score E
Sequences producing significant alignments: (bits) Value
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 284 5e-77
AL366725 274 4e-74
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 58 7e-09
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 49 3e-06
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 49 4e-06
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 48 6e-06
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 48 8e-06
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 47 2e-05
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 43 2e-04
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 36 0.030
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 34 0.086
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 32 0.56
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 31 0.73
TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis t... 31 0.73
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 31 0.95
TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein... 30 2.1
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 29 2.8
TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine... 29 2.8
TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein cons... 29 2.8
BE997729 29 3.6
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 284 bits (726), Expect = 5e-77
Identities = 138/171 (80%), Positives = 149/171 (86%), Gaps = 2/171 (1%)
Frame = +2
Query: 1 MAGRNDAAIAAALEAVAQAVQQQPQAGNGE--VRMLETFLRNHPPAFKGRYDPDGAQSWL 58
M G +DAA+ AALEAVAQAVQQ P+ G RMLETFLRNHPP FKGRY PDGA WL
Sbjct: 2 MTGSSDAALVAALEAVAQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWL 181
Query: 59 KEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRREFLNRY 118
KE+ERIFRVMQC E QKV+FGTHMLAEEADDWW+SLLPVLEQ+ AVVTWA+FR+EFL RY
Sbjct: 182 KEIERIFRVMQCFETQKVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRY 361
Query: 119 FPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYPHYAPETAEFSKCI 169
FPEDVRG+KEIEFLELKQGDMSV EYAAKFVELA FYPHY+ ETAEFSKCI
Sbjct: 362 FPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELATFYPHYSAETAEFSKCI 514
>AL366725
Length = 485
Score = 274 bits (701), Expect = 4e-74
Identities = 123/161 (76%), Positives = 141/161 (87%)
Frame = +2
Query: 153 KFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYKVM 212
KFYPHYA ETAEFSKCIKFENGLR DIKRAIGYQQ+R+F DLV+ CRIYEEDTKAH KV+
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 213 SERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDE 272
+ER+ KG +RPKPYSAP DKGKQR+ D+RRP K+DAPAEIVCF GEKGHKSNVC K+
Sbjct: 182 NERKTKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEI 361
Query: 273 KKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
KKC RC +KGH++ADCKR D+VC+NCNEEGHI +QC QPK+
Sbjct: 362 KKCVRCDKKGHIVADCKRNDIVCFNCNEEGHIGSQCKQPKR 484
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 57.8 bits (138), Expect = 7e-09
Identities = 34/123 (27%), Positives = 60/123 (48%), Gaps = 5/123 (4%)
Frame = +1
Query: 196 SRCRIYEEDTKAHYKVMSERRGKGHQNRPKPYSAPTDKGKQRLN---DERRPSK--RDAP 250
SR R + +++ V R + +R PY + +G + N + +RP R+ P
Sbjct: 289 SRSRSPMDRSRSRSPVDRRIRSERFSHREAPYRRDSRRGFSQDNLCKNCKRPGHYVRECP 468
Query: 251 AEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
VC C GH ++ C+ + C+ C + GHM + C + +C+ C + GH + +CT
Sbjct: 469 NVAVCHNCSLPGHIASECST-KSLCWNCKEPGHMASSCPN-EGICHTCGKAGHRARECTV 642
Query: 311 PKK 313
P+K
Sbjct: 643 PQK 651
Score = 51.2 bits (121), Expect = 7e-07
Identities = 24/65 (36%), Positives = 37/65 (56%), Gaps = 6/65 (9%)
Frame = +1
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCK-----RGDV-VCYNCNEEGHISTQ 307
+C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC ++GHI+ +
Sbjct: 535 LCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVE 711
Query: 308 CTQPK 312
CT K
Sbjct: 712 CTNEK 726
Score = 50.8 bits (120), Expect = 9e-07
Identities = 25/66 (37%), Positives = 37/66 (55%)
Frame = +1
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
+C C ++GH + CT +EK C C + GH+ DC D +C CN GH++ QC PK
Sbjct: 670 LCNNCYKQGHIAVECT-NEKACNNCRKTGHLARDCPN-DPICNLCNISGHVARQC--PKS 837
Query: 314 VRVGGK 319
+G +
Sbjct: 838 NVIGDR 855
Score = 50.4 bits (119), Expect = 1e-06
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 6/65 (9%)
Frame = +1
Query: 250 PAEIVCFKCGEKGHKSNVCTKDEKK------CFRCGQKGHMLADCKRGDVVCYNCNEEGH 303
P E +C CG+ GH++ CT +K C C ++GH+ +C + C NC + GH
Sbjct: 580 PNEGICHTCGKAGHRARECTVPQKPPGDLRLCNNCYKQGHIAVEC-TNEKACNNCRKTGH 756
Query: 304 ISTQC 308
++ C
Sbjct: 757 LARDC 771
Score = 42.4 bits (98), Expect = 3e-04
Identities = 24/85 (28%), Positives = 32/85 (37%), Gaps = 23/85 (27%)
Frame = +1
Query: 247 RDAPAEIVCFKCGEKGHKSNVCTKD----------------------EKKCFRCGQKGHM 284
RD P + +C C GH + C K + C C Q GHM
Sbjct: 763 RDCPNDPICNLCNISGHVARQCPKSNVIGDRGGGGSLRGGYRDGGFRDVVCRSCQQFGHM 942
Query: 285 LADCKRGD-VVCYNCNEEGHISTQC 308
DC G ++C NC GH + +C
Sbjct: 943 SRDCMGGPLMICQNCGGRGHQAYEC 1017
Score = 32.3 bits (72), Expect = 0.33
Identities = 15/44 (34%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Frame = +1
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK-CFRCGQKGHMLADCKRGDVV 294
++VC C + GH S C C CG +GH +C G V
Sbjct: 904 DVVCRSCQQFGHMSRDCMGGPLMICQNCGGRGHQAYECPSGRFV 1035
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 48.9 bits (115), Expect = 3e-06
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 2/80 (2%)
Frame = -2
Query: 43 PAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNG 102
P F+G PD WL+ +ER+F+ + E QKV+ L + A WW ++ ++ G
Sbjct: 724 PDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREG 545
Query: 103 --AVVTWAVFRREFLNRYFP 120
+ TW R++ +Y P
Sbjct: 544 KSKIKTWEKMRQKLTRKYLP 485
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 48.5 bits (114), Expect = 4e-06
Identities = 22/44 (50%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Frame = +2
Query: 274 KCFRCGQKGHMLADCKRGD--VVCYNCNEEGHISTQC-TQPKKV 314
+CF CG GH DCK GD CY C E GHI C PKK+
Sbjct: 356 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKL 487
Score = 42.4 bits (98), Expect = 3e-04
Identities = 22/64 (34%), Positives = 32/64 (49%), Gaps = 7/64 (10%)
Frame = +2
Query: 233 KGKQRLNDERRPSKR-----DAPAEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHML 285
KG R + + R S+ P CF CG GH + C + KC+RCG++GH+
Sbjct: 278 KGVPRGSRDSRDSREYLGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIE 457
Query: 286 ADCK 289
+CK
Sbjct: 458 KNCK 469
Score = 28.9 bits (63), Expect = 3.6
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +2
Query: 255 CFKCGEKGHKSNVCTKDEKKCFR 277
C++CGE+GH C KK R
Sbjct: 425 CYRCGERGHIEKNCKNSPKKLSR 493
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 48.1 bits (113), Expect = 6e-06
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 2/78 (2%)
Frame = +2
Query: 43 PAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPV--LEQ 100
P F G+ PD WL+ +ER+F+ + +E QKV+ L + A WW +L E
Sbjct: 347 PDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKVKIVAAKLKKHASIWWKNLKRKRNCEG 526
Query: 101 NGAVVTWAVFRREFLNRY 118
+ TW R++ +Y
Sbjct: 527 KSKIKTWDKMRQKLTRKY 580
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 47.8 bits (112), Expect = 8e-06
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 12/67 (17%)
Frame = +3
Query: 254 VCFKCGEKGHKSNVCT-----KDEKKCFRCGQKGHMLADC----KRGDVV---CYNCNEE 301
+C +C +GH++ C +D K + CG GH LA+C + G + C+ C E+
Sbjct: 354 ICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQ 533
Query: 302 GHISTQC 308
GH+S C
Sbjct: 534 GHLSKNC 554
Score = 44.7 bits (104), Expect = 6e-05
Identities = 32/121 (26%), Positives = 48/121 (39%), Gaps = 22/121 (18%)
Frame = +3
Query: 224 PKPYSAPTDKGKQRLNDER-----RPSKRDAPAEIV-------CFKCGEKGHKSNVCTKD 271
P P + P+ K K + ++ +P P + CF C H + CT+
Sbjct: 153 PTPPNDPSKKKKNKFKRKKPDSNSKPRTGKRPLRVPGMKPGDSCFICKGLDHIAKFCTQK 332
Query: 272 -----EKKCFRCGQKGHMLADCKRGD-----VVCYNCNEEGHISTQCTQPKKVRVGGKVF 321
K C RC ++GH +C G YNC + GH C P ++ GG +F
Sbjct: 333 AEWEKNKICLRCRRRGHRAQNCPDGGSKEDFKY*YNCGDNGHSLANCPHP--LQEGGTMF 506
Query: 322 A 322
A
Sbjct: 507 A 509
Score = 27.7 bits (60), Expect = 8.1
Identities = 11/42 (26%), Positives = 20/42 (47%), Gaps = 7/42 (16%)
Frame = +3
Query: 256 FKCGEKGHKSNVCTKDEKK-------CFRCGQKGHMLADCKR 290
+ CG+ GH C ++ CF C ++GH+ +C +
Sbjct: 435 YNCGDNGHSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNCPK 560
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Frame = +2
Query: 43 PAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNG 102
P F+G D WL+ +ER+F + E QKV+ L + A WW +L ++ G
Sbjct: 629 PDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREG 808
Query: 103 --AVVTWAVFRREFLNRYFP 120
+ TW R++ +Y P
Sbjct: 809 KSKIKTWDKMRQKLTRKYLP 868
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 43.1 bits (100), Expect = 2e-04
Identities = 22/88 (25%), Positives = 30/88 (34%), Gaps = 31/88 (35%)
Frame = +3
Query: 255 CFKCGEKGHKSNVCTKDEKK-----------------CFRCGQKGHMLADCKRGD----- 292
C+ CG+ GH + C + ++ C+ CG H DC RG
Sbjct: 351 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 530
Query: 293 ---------VVCYNCNEEGHISTQCTQP 311
CY C GHI+ C P
Sbjct: 531 GGGGYGGGGTSCYRCGGVGHIARDCATP 614
Score = 28.5 bits (62), Expect = 4.7
Identities = 15/71 (21%), Positives = 25/71 (35%), Gaps = 14/71 (19%)
Frame = +3
Query: 232 DKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDE--------------KKCFR 277
D+ + ++R + C+ CG H + C + C+R
Sbjct: 393 DRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYR 572
Query: 278 CGQKGHMLADC 288
CG GH+ DC
Sbjct: 573 CGGVGHIARDC 605
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 35.8 bits (81), Expect = 0.030
Identities = 16/57 (28%), Positives = 20/57 (35%), Gaps = 20/57 (35%)
Frame = +1
Query: 255 CFKCGEKGHKSNVCTKDEK--------------------KCFRCGQKGHMLADCKRG 291
C+ CGE GH + CT C+ CG+ GH DC G
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 34.7 bits (78), Expect = 0.066
Identities = 15/54 (27%), Positives = 20/54 (36%), Gaps = 20/54 (37%)
Frame = +1
Query: 275 CFRCGQKGHMLADCKR--------------------GDVVCYNCNEEGHISTQC 308
C+ CG+ GHM +C G CY+C E GH + C
Sbjct: 16 CYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
Score = 30.4 bits (67), Expect = 1.2
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +1
Query: 295 CYNCNEEGHISTQCT 309
CYNC E GH++ +CT
Sbjct: 16 CYNCGESGHMARECT 60
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 34.3 bits (77), Expect = 0.086
Identities = 14/48 (29%), Positives = 20/48 (41%), Gaps = 13/48 (27%)
Frame = -1
Query: 274 KCFRCGQKGHMLADCKRGDVV-------------CYNCNEEGHISTQC 308
KC++C Q GH ++C CY CN+ GH + C
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNC 361
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 31.6 bits (70), Expect = 0.56
Identities = 14/29 (48%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Frame = -1
Query: 295 CYNCNEEGHISTQCTQ---PKKVRVGGKV 320
CY+C+E GHI+ CT KK + G KV
Sbjct: 316 CYSCHERGHIARNCTNTSAAKKKKKGPKV 230
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 31.2 bits (69), Expect = 0.73
Identities = 21/81 (25%), Positives = 38/81 (45%), Gaps = 6/81 (7%)
Frame = +1
Query: 215 RRGKGHQNRPKPYSAPTDKGKQRLN-----DERRPSKRDAPAEIVCFKCGEKGHKSNVCT 269
R KG + K A D+ ++ + +P K++ ++ + +KG KS+
Sbjct: 886 RGQKGKLKKMKEKYADQDEEERSIRMSLLASSGKPIKKEETLPVI--ETSDKGKKSDSGP 1059
Query: 270 KDEKK-CFRCGQKGHMLADCK 289
D K C++C + GH+ DCK
Sbjct: 1060IDAPKICYKCKKVGHLSRDCK 1122
>TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis thaliana},
partial (23%)
Length = 727
Score = 31.2 bits (69), Expect = 0.73
Identities = 9/18 (50%), Positives = 15/18 (83%)
Frame = +2
Query: 272 EKKCFRCGQKGHMLADCK 289
++KC+ CGQ GH+ A+C+
Sbjct: 200 QEKCYMCGQVGHLAAECR 253
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 30.8 bits (68), Expect = 0.95
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 2/20 (10%)
Frame = +1
Query: 275 CFRCGQKGHMLADC--KRGD 292
CF CG+ GH +DC KRGD
Sbjct: 166 CFTCGESGHRASDCPNKRGD 225
Score = 28.9 bits (63), Expect = 3.6
Identities = 20/64 (31%), Positives = 28/64 (43%), Gaps = 8/64 (12%)
Frame = +1
Query: 213 SERRG----KGHQNRPKPYSAPTDK----GKQRLNDERRPSKRDAPAEIVCFKCGEKGHK 264
S RRG K + KP + D G+Q P++ A CF CGE GH+
Sbjct: 25 SSRRGGRSYKSGNSWSKPERSSRDDWLIGGRQSSRSSSSPNRSFAGT---CFTCGESGHR 195
Query: 265 SNVC 268
++ C
Sbjct: 196 ASDC 207
>TC81162 weakly similar to PIR|T47673|T47673 hypothetical protein T26I12.220
- Arabidopsis thaliana, partial (9%)
Length = 756
Score = 29.6 bits (65), Expect = 2.1
Identities = 10/28 (35%), Positives = 18/28 (63%)
Frame = +2
Query: 285 LADCKRGDVVCYNCNEEGHISTQCTQPK 312
+A KR + +CY C ++GH ++C P+
Sbjct: 392 VASGKRKNRMCYGCRQKGHNLSECPNPQ 475
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 29.3 bits (64), Expect = 2.8
Identities = 8/19 (42%), Positives = 13/19 (68%)
Frame = +1
Query: 271 DEKKCFRCGQKGHMLADCK 289
D+ KC+ CG+ GH +C+
Sbjct: 307 DDLKCYECGEPGHFARECR 363
>TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine zipper
protein HDZ3 {Phaseolus vulgaris}, complete
Length = 1532
Score = 29.3 bits (64), Expect = 2.8
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 3/45 (6%)
Frame = +2
Query: 198 CRIYEEDTKAHYKVMSERRGKGHQNRP---KPYSAPTDKGKQRLN 239
CR+ T H+K E GHQ P KP+S+ + K N
Sbjct: 11 CRLSHTHTHTHHKTSPENSISGHQRSPSSGKPFSSKLNPFKDHRN 145
>TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein constans-like
14. [Mouse-ear cress] {Arabidopsis thaliana}, partial
(27%)
Length = 937
Score = 29.3 bits (64), Expect = 2.8
Identities = 16/77 (20%), Positives = 32/77 (40%), Gaps = 3/77 (3%)
Frame = +1
Query: 240 DERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEKK---CFRCGQKGHMLADCKRGDVVCY 296
D+R + A +C C H +N + + C RC + ++ + +C
Sbjct: 265 DQRSMVYCRSDAACLCLSCDRNVHSANTLARRHSRTLLCERCSSQPALVRCSEEKVSLCQ 444
Query: 297 NCNEEGHISTQCTQPKK 313
NC+ GH ++ + K+
Sbjct: 445 NCDWLGHGNSTSSNHKR 495
>BE997729
Length = 559
Score = 28.9 bits (63), Expect = 3.6
Identities = 16/48 (33%), Positives = 20/48 (41%)
Frame = -2
Query: 263 HKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
HK +C+ + CF H L Y C+EEG I T C Q
Sbjct: 315 HKEGLCSIWIRHCFLSDPHAHFLLSPS------YCCSEEGEIETICYQ 190
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,919,108
Number of Sequences: 36976
Number of extensions: 192830
Number of successful extensions: 905
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 873
length of query: 396
length of database: 9,014,727
effective HSP length: 98
effective length of query: 298
effective length of database: 5,391,079
effective search space: 1606541542
effective search space used: 1606541542
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136954.7


