

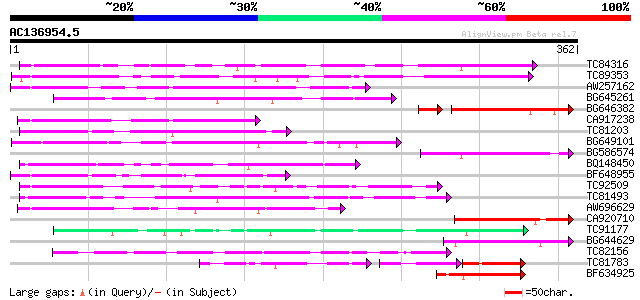
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.5 - phase: 0
(362 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 123 1e-28
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 117 9e-27
AW257162 111 5e-25
BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter p... 102 2e-22
BG646382 88 3e-20
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 92 2e-19
TC81203 weakly similar to GP|10177464|dbj|BAB10855. gb|AAD21700.... 91 7e-19
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 89 3e-18
BG586574 84 7e-17
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 84 1e-16
BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3... 79 3e-15
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 76 2e-14
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 76 2e-14
AW696629 75 4e-14
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 72 3e-13
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 68 5e-12
BG644629 67 8e-12
TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein famil... 66 2e-11
TC81783 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 39 2e-11
BF634925 62 5e-10
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 123 bits (308), Expect = 1e-28
Identities = 101/336 (30%), Positives = 157/336 (46%), Gaps = 5/336 (1%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP ELI ILLR V SL+ K V K W TL SD F H +S L +A S
Sbjct: 42 LPEELIVI-ILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQL-VACESVS 215
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGL 126
R ++ + P SLL S+T+ IP+++ +++ I+GSCNG
Sbjct: 216 AYR----------TWEIKTYPIE-SLLENSSTTVIPVSNTGHQRYT------ILGSCNGF 344
Query: 127 ICLHGCFHGSGYKKHSFC--FWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDTYKTVM 184
+CL Y + C WNP+ KSK+ P+ + GFGYD YK +
Sbjct: 345 LCL--------YDNYQRCVRLWNPSINLKSKSS---PTIDRFIYYGFGYDQVNHKYKLLA 491
Query: 185 FGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQ-SSFPVELALRSRWDDIKYDGVYLSNS 243
R ++T G++ ++++ FP R K+ G ++S +
Sbjct: 492 VKAFS---------RITETMIYTFGENSCKNVEVKDFPRYPPNR------KHLGKFVSGT 626
Query: 244 ISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFD--NPNICALMDCICFS 301
++W+V R + I+S D+E ETY Q+ LP+ + +P + L +CIC
Sbjct: 627 LNWIVDER---------DGRATILSFDIEKETYRQVLLPQHGYAVYSPGLYVLSNCICVC 779
Query: 302 YDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGIN 337
F +T + +W MK++GV ESWT+ + I ++NL I+
Sbjct: 780 TSFLDTRWQLWMMKKYGVAESWTKLMSIPHENLFIS 887
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 117 bits (292), Expect = 9e-27
Identities = 110/347 (31%), Positives = 159/347 (45%), Gaps = 14/347 (4%)
Frame = +3
Query: 2 NISEI---LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNL 58
++SEI +P E+I EILLR V SL+ + V K W TLISDP F K H+ +S + L
Sbjct: 81 SVSEITADMPEEIIV-EILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQL 257
Query: 59 RLALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCG 118
S L SY + LL + + AD ++
Sbjct: 258 VSVFVSIAKCNL--------VSYPLK------PLLDNPSAHRVEPAD---FEMIHTTSMT 386
Query: 119 IIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSK---TLLYVPSYLNRVRL--GFGY 173
IIGSCNGL+CL + F WNP+ + KSK T++ S+ ++ L GFGY
Sbjct: 387 IIGSCNGLLCLSDFYQ--------FTLWNPSIKLKSKPSPTIIAFDSFDSKRFLYRGFGY 542
Query: 174 DNSTDTYKT---VMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRW 230
D D YK V +DE ++T G W IQ FP + SR
Sbjct: 543 DQVNDRYKVLAVVQNCYNLDE---------TKTLIYTFGGKDWTTIQK-FPCD---PSRC 683
Query: 231 DDIKYD-GVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFD-N 288
D + G ++S +++W+V + VI+ D+E ETY ++ LP+ D N
Sbjct: 684 DLGRLGVGKFVSGNLNWIVSKK-------------VIVFFDIEKETYGEMSLPQDYGDKN 824
Query: 289 PNICALMDCICFSYDFK-ETHFVIWQMKEFGVEESWTQFLKISYQNL 334
+ + I S+D +TH+V+W MKE+GV ESWT+ + I L
Sbjct: 825 TVLYVSSNRIYVSFDHSNKTHWVVWMMKEYGVVESWTKLMIIPQDKL 965
>AW257162
Length = 728
Score = 111 bits (277), Expect = 5e-25
Identities = 84/231 (36%), Positives = 112/231 (48%), Gaps = 1/231 (0%)
Frame = +1
Query: 1 MNISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRL 60
++ S + P ++I EIL V +LM +K VSK WNTLISD FVKMHL R
Sbjct: 109 LSSSPVFPDDIIA-EILSWLTVKTLMKMKCVSKSWNTLISDSNFVKMHLN--------RS 261
Query: 61 ALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGII 120
A S L + R P SV L+ + ++P D YQ DC G++
Sbjct: 262 ARHSQSYLVSEHRG-------DYNFVPFSVRGLMNGRSITLP--KDPYYQLIEKDCPGVV 414
Query: 121 GSCNGLICLHGCFHG-SGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDT 179
GSCNGL+CL GC +++ WNPATR+ S L + + L FGYDN+T T
Sbjct: 415 GSCNGLVCLSGCVADVEEFEEMWLRIWNPATRTISDKLYFSANRLQCWEFMFGYDNTTQT 594
Query: 180 YKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRW 230
YK V L + T V + ++IWR+IQ SFP E + W
Sbjct: 595 YKVV--------ALYPDSEMTTKVGIICFRNNIWRNIQ-SFPCEASSVLYW 720
>BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter permease
protein SSO1032 [imported] - Sulfolobus solfataricus,
partial (4%)
Length = 771
Score = 102 bits (254), Expect = 2e-22
Identities = 78/234 (33%), Positives = 113/234 (47%), Gaps = 15/234 (6%)
Frame = +1
Query: 29 KFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQIRAGGRGCSYTVTVAPT 88
+F++ W ++I P FVK+HL S K +L + R+ + C+ + TV
Sbjct: 112 RFINILWKSIIDHPTFVKLHLNRSSQKPDLTFV----SSARITLWTLESTCTVSFTVFR- 276
Query: 89 SVSLLLESTTSSIPIADD-LQYQFSCVDCCGIIGSCNGLICLHG--CFHGSGYKKHSFCF 145
LLE+ I ++DD YQ DC IIGSCNGL+CL G G+ S F
Sbjct: 277 ----LLENPPVIINLSDDDPYYQLKDKDCVHIIGSCNGLLCLFGLKINDSCGHMDISIRF 444
Query: 146 WNPATRSKSKTLLYVPSYLNR-----------VRLGFGYDNSTDTYKTVMFGITMDEGLG 194
WNPA+R SK + Y +N ++ FGY NS+DTYK V F +
Sbjct: 445 WNPASRKISKKVGYCGDGVNNDPSCFPGKGDLLKFVFGYVNSSDTYKVVYFIV------- 603
Query: 195 GNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNS-ISWL 247
T +VF+LGD++WR I++S PV + S + + G + NS + WL
Sbjct: 604 ----GTTSARVFSLGDNVWRKIENS-PVVIHHVSGFVHLSGSGFLVDNSELPWL 750
>BG646382
Length = 603
Score = 87.8 bits (216), Expect(2) = 3e-20
Identities = 46/88 (52%), Positives = 58/88 (65%), Gaps = 10/88 (11%)
Frame = +3
Query: 283 KLPFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISY-----QNLGIN 337
++P P++ LMDC+CFS+DFK T FVIWQMKEFG +ESWTQ +I Y NL IN
Sbjct: 213 EVPCVEPSLRVLMDCLCFSHDFKRTEFVIWQMKEFGSQESWTQLFRIKYINLQIHNLPIN 392
Query: 338 YILGRNGFL-----VLPVCLSENGATLI 360
L G++ +LP+ LSENG TLI
Sbjct: 393 DNLDLLGYMECNIPLLPLYLSENGDTLI 476
Score = 28.1 bits (61), Expect(2) = 3e-20
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +2
Query: 262 EQFVIISLDLETETY 276
EQ VI+SLDL TETY
Sbjct: 137 EQLVIVSLDLSTETY 181
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 92.4 bits (228), Expect = 2e-19
Identities = 59/155 (38%), Positives = 83/155 (53%)
Frame = +2
Query: 6 ILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSN 65
+LP ELIT E+L R DV SLM ++ VS+ WN++ISDP FVK+H+K S +L L+L +
Sbjct: 143 VLPDELIT-EVLSRGDVKSLMRMRCVSEYWNSMISDPRFVKLHMKRSARNAHLTLSLCKS 319
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNG 125
G V P V L+E+ ++P D Y+ +C +IGSCNG
Sbjct: 320 ------------GIDGDNNVVPYPVRGLIENGLITLP--SDPYYRLKDKECQYVIGSCNG 457
Query: 126 LICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYV 160
+CL G Y+ F FWNPA ++ L Y+
Sbjct: 458 WLCLLGFSSIGAYRHIWFRFWNPAMGKMTQKLGYI 562
>TC81203 weakly similar to GP|10177464|dbj|BAB10855.
gb|AAD21700.1~gene_id:MQB2.18~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 715
Score = 90.9 bits (224), Expect = 7e-19
Identities = 70/187 (37%), Positives = 92/187 (48%), Gaps = 13/187 (6%)
Frame = +1
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLIS-DPIFVKMHLKLSKSKGNLRLALFSN 65
L ++ + +IL R V +LM K V K W TLIS DP F K+HL+ +S N L L S+
Sbjct: 196 LLLDELIVDILSRLPVKTLMQFKCVCKSWKTLISHDPSFAKLHLQ--RSPRNTHLTLVSD 369
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIP-----------IADDLQYQFSCV 114
++ +V P VS L+E+ + P I DD Y +
Sbjct: 370 RS---------SDDESNFSVVPFPVSHLIEAPLTITPFEPYHLLRNVAIPDDPYYVLGNM 522
Query: 115 DCCGIIGSCNGLICLHGCFHGSGYKKHS-FCFWNPATRSKSKTLLYVPSYLNRVRLGFGY 173
DCC IIGSCNGL CL ++H F FWNPAT + S+ L + + RL FGY
Sbjct: 523 DCCIIIGSCNGLYCLRCYSLIYEEEEHDWFRFWNPATNTLSEELGCLNEFF---RLTFGY 693
Query: 174 DNSTDTY 180
D S DTY
Sbjct: 694 DISNDTY 714
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 88.6 bits (218), Expect = 3e-18
Identities = 81/261 (31%), Positives = 112/261 (42%), Gaps = 12/261 (4%)
Frame = +1
Query: 2 NISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLA 61
++ ++P +LI + P V ++ LK VSK WNTLI+ P F+K+HL S N L
Sbjct: 70 SVPVVIPSDLIAIILTFLP-VKTITQLKLVSKSWNTLITSPSFIKIHLNQSSQNPNFILT 246
Query: 62 LFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIG 121
S K + V + LL T S D + ++G
Sbjct: 247 P-SRKQYSIN------------NVLSVPIPRLLTGNTVS---GDTYHNILNNDHHFRVVG 378
Query: 122 SCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTL----LYVPSYLNRVRLGFGYDNST 177
SCNGL+CL + K F WNPATR+ S+ L Y P + R FG D
Sbjct: 379 SCNGLLCLLFKSEFITHLKFRFRIWNPATRTISEELGFFRKYKPLFGGVSRFTFGCDYLR 558
Query: 178 DTYKTVMFGITMDEGLGGNRMRTAVVKVFTLG----DSIWRDIQSSF----PVELALRSR 229
TYK V D G+ MR+ V+VF LG D WR+I + F V L+
Sbjct: 559 GTYKLVALHTVED----GDVMRSN-VRVFNLGNDDSDKCWRNIPNPFVCADGVHLSGTGN 723
Query: 230 WDDIKYDGVYLSNSISWLVCH 250
W ++ D Y+ S+ L H
Sbjct: 724 WLSLREDARYIEGSMEPLTPH 786
>BG586574
Length = 732
Score = 84.3 bits (207), Expect = 7e-17
Identities = 50/109 (45%), Positives = 60/109 (54%), Gaps = 11/109 (10%)
Frame = +1
Query: 263 QFVIISLDLETETYTQLQLPKLPFD-----------NPNICALMDCICFSYDFKETHFVI 311
Q VIISLDL TETYTQ P + D P + LMD +CF +D ET FVI
Sbjct: 55 QLVIISLDLGTETYTQFLPPPISLDLSHVLQKVSHAKPGVSMLMDSLCFYHDLNETDFVI 234
Query: 312 WQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCLSENGATLI 360
W+M +FG E+SW Q LKISY L +N G + F L NG TL+
Sbjct: 235 WKMTKFGDEKSWAQLLKISYHKLKMNLKPGISMF-----NLYVNGDTLV 366
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 83.6 bits (205), Expect = 1e-16
Identities = 74/222 (33%), Positives = 111/222 (49%), Gaps = 4/222 (1%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP E I EIL R V LM + V K W + IS P FVK HL++S ++ +L L FS
Sbjct: 3 LPFE-IQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTR-HLFLLTFSKL 176
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVD-CCGIIGSCNG 125
+ L I+ SY ++ S+ E T P L+Y + D ++GSC+G
Sbjct: 177 SPELVIK------SYPLS------SVFTEMT----PTFTQLEYPLNNRDESDSMVGSCHG 308
Query: 126 LICLHGCFHGSGYKKHSF-CFWNPATR--SKSKTLLYVPSYLNRVRLGFGYDNSTDTYKT 182
++C+ C SF WNP+ R +K + + + FGYD+S+DTYK
Sbjct: 309 ILCIQ-C-------NLSFPVLWNPSIRKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKV 464
Query: 183 VMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVEL 224
V T + G +++T +V V T+G + WR IQ+ FP ++
Sbjct: 465 VAVFCTSNIDNGVYQLKT-LVNVHTMGTNCWRRIQTEFPFKI 587
>BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3661715) {Mus
musculus}, partial (3%)
Length = 514
Score = 79.0 bits (193), Expect = 3e-15
Identities = 69/179 (38%), Positives = 88/179 (48%)
Frame = +3
Query: 1 MNISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRL 60
M + LP ELI EI+ V LM + VSK + TLISDP FV+MHL+ KS N L
Sbjct: 39 MASAAFLPSELIV-EIISWLPVKYLMQFRCVSKFYKTLISDPYFVQMHLE--KSARNPHL 209
Query: 61 ALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGII 120
AL +L +R G ++ SVS LL + ++ P Y SC II
Sbjct: 210 ALMWQDDL---LREDG-------SIIFLSVSRLLGNKYTTPPFQSGTFYH-SC-----II 341
Query: 121 GSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDT 179
GSCNGL+CL Y + FWNPATR+K +L S + FGYD + T
Sbjct: 342 GSCNGLLCLVDFHCPDYYYYYYLYFWNPATRTKFXNILITLS--XDFKFSFGYDTLSKT 512
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 76.3 bits (186), Expect = 2e-14
Identities = 84/283 (29%), Positives = 124/283 (43%), Gaps = 13/283 (4%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP EL+ EIL R V L L+ + K +N+LISDP F K HL S + +L L
Sbjct: 174 LPFELVA-EILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTPHHLIL------ 332
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV--------DCCG 118
R+ + + V+P L +TS++P+ +C+ + C
Sbjct: 333 ------RSNNGSGRFALIVSPIQSVL----STSTVPVPQTQLTYPTCLTEEFASPYEWC- 479
Query: 119 IIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVR-----LGFGY 173
SC+G+ICL + S WNP +K KTL + Y++ R FGY
Sbjct: 480 ---SCDGIICLTTDY-------SSAVLWNPFI-NKFKTLPPL-KYISLKRSPSCLFTFGY 623
Query: 174 DNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDI 233
D D YK +F IT ++ V+V T+G S WR I+ FP W I
Sbjct: 624 DPFADNYK--VFAITF-------CVKRTTVEVHTMGTSSWRRIE-DFP-------SWSFI 752
Query: 234 KYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETY 276
G++++ + WL Q++ I+SLDLE E+Y
Sbjct: 753 PDSGIFVAGYVHWLTYDGPGSQRE--------IVSLDLEDESY 857
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 75.9 bits (185), Expect = 2e-14
Identities = 73/282 (25%), Positives = 119/282 (41%), Gaps = 6/282 (2%)
Frame = +1
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP E +TTEIL R L+ L+ K W LI F+ +HL SKS+ ++ +
Sbjct: 16 LPTE-VTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHL--SKSRDSVII------ 168
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV-DCCGIIGSCNG 125
LR R L E +S+ +L + C + ++GSCNG
Sbjct: 169 -LRQHSR-------------------LYELDLNSMDRVKELDHPLMCYSNRIKVLGSCNG 288
Query: 126 LICLHG-----CFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDTY 180
L+C+ F +KH P R ++ + + L GFGYD++TD Y
Sbjct: 289 LLCICNIADDIAFWNPTIRKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDY 468
Query: 181 KTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYL 240
K V +D NR + V ++T+G +W + P L + GV++
Sbjct: 469 KLVSISNFVDL---HNRSYDSHVTIYTMGSDVWMPL-PGVPYALCC------ARPMGVFV 618
Query: 241 SNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP 282
S ++ W+V + ++L I++ DL E + ++ LP
Sbjct: 619 SGALHWVVPRALEPDSRDL------IVAFDLRFEVFREVALP 726
>AW696629
Length = 663
Score = 75.1 bits (183), Expect = 4e-14
Identities = 74/220 (33%), Positives = 99/220 (44%), Gaps = 11/220 (5%)
Frame = +2
Query: 6 ILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSN 65
ILP +LI +IL V L+ VSK W +LI DP F K+HL+ S ++ L +
Sbjct: 62 ILPSDLIM-QILSWLPVKLLIRFTSVSKHWKSLILDPNFAKLHLQKSPKNTHMILTALDD 238
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCC-------G 118
++ ++ VT P SLLLE Q FS +CC
Sbjct: 239 ED-----------DTWVVTPYPVR-SLLLE------------QSSFSDEECCCFDYHSYF 346
Query: 119 IIGSCNGLICLHGCFHGSGYKKHSFC-FWNPATRSKSKTL--LYVPSYLNRVRLGFGYDN 175
I+GS NGL+CL K F FWNP+ R +SK L + Y RLGFGYD+
Sbjct: 347 IVGSTNGLVCLAVEKSLENRKYELFIKFWNPSLRLRSKKAPSLNIGLY-GTARLGFGYDD 523
Query: 176 STDTYKTV-MFGITMDEGLGGNRMRTAVVKVFTLGDSIWR 214
DTYK V +F + G +V +GDS W+
Sbjct: 524 LNDTYKAVAVFWDHTTHKMEG--------RVHCMGDSCWK 619
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 72.0 bits (175), Expect = 3e-13
Identities = 36/83 (43%), Positives = 51/83 (61%), Gaps = 7/83 (8%)
Frame = -2
Query: 285 PFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNL-------GIN 337
P P + L+DC+CF +D +T FVIWQMKEFGV ESWT+ KI Y +L +
Sbjct: 773 PCAEPYLRVLLDCLCFLHDLGKTEFVIWQMKEFGVRESWTRLFKIPYVDLQMLNLPIDVQ 594
Query: 338 YILGRNGFLVLPVCLSENGATLI 360
Y+ N + +LP+ +S+NG +I
Sbjct: 593 YL---NEYPMLPLYISKNGDKVI 534
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 68.2 bits (165), Expect = 5e-12
Identities = 78/320 (24%), Positives = 129/320 (39%), Gaps = 17/320 (5%)
Frame = +2
Query: 29 KFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFS----NKNLRLQIRAGGRGCSYTVT 84
K V K W +LIS P F H +L+ + R+ L + + ++ L++ YT
Sbjct: 77 KCVCKLWLSLISQPHFANSHFQLTTATHTNRIMLITPYLQSLSIDLELSLNDDSAVYT-- 250
Query: 85 VAPTSVSLLLEST---TSSIPIADDLQ--YQFSCVDCCGIIGSCNGLICLHGCFHGSGYK 139
T +S L++ +SS DDL F +D GSC G I L+ C+
Sbjct: 251 ---TDISFLIDDEDYYSSSSSDMDDLSPPKSFFILD---FKGSCRGFILLN-CYS----- 394
Query: 140 KHSFCFWNPATRSKSKTLLYVPSYLN---RVRLGFGYDNSTDTYKTVMFGITMDEGLGGN 196
S C WNP+T K + + N GFGYD STD Y + G
Sbjct: 395 --SLCIWNPST-GFHKRIPFTTIDSNPDANYFYGFGYDESTDDYLVISMSYEPSPSSDG- 562
Query: 197 RMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQQ 256
+ + +F+L ++W ++ L L S+ + + +I WL
Sbjct: 563 --MLSHLGIFSLRANVWTRVEGG---NLLLYSQNSLLNLVESLSNGAIHWLAFRN----- 712
Query: 257 KNLTTEQFVIISLDLETETYTQLQLPKLPFDNPN----ICALMDCICFSYDFKE-THFVI 311
VI++ L +L+LP + P+ + C+ + + F I
Sbjct: 713 ---DISMPVIVAFHLMERKLLELRLPNEIINGPSRAYDLWVYRGCLALWHILPDRVTFQI 883
Query: 312 WQMKEFGVEESWTQFLKISY 331
W M+++ V+ SWT+ L +S+
Sbjct: 884 WVMEKYNVQSSWTKTLVLSF 943
>BG644629
Length = 546
Score = 67.4 bits (163), Expect = 8e-12
Identities = 40/97 (41%), Positives = 54/97 (55%), Gaps = 14/97 (14%)
Frame = -3
Query: 278 QLQLPK----LPFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQN 333
Q +LP+ +P P + +C SY +KET F+IWQMKE +ESWTQFLKI YQN
Sbjct: 544 QYRLPRDFHEMPSALPIVAV*GGFLCCSYFYKETDFLIWQMKELANDESWTQFLKIRYQN 365
Query: 334 LGIN----------YILGRNGFLVLPVCLSENGATLI 360
L IN + + F ++P+ LSE TL+
Sbjct: 364 LQINHDYFSDHEFIHDTVKYHFQLVPLLLSEEADTLV 254
>TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein family AtFBX9
{Arabidopsis thaliana}, partial (4%)
Length = 1136
Score = 66.2 bits (160), Expect = 2e-11
Identities = 70/256 (27%), Positives = 110/256 (42%), Gaps = 1/256 (0%)
Frame = +3
Query: 28 LKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQIRAGGRGCSYTVTVAP 87
+K VSK W +LISD F +K NLR++ S++ L ++ G Y
Sbjct: 456 VKSVSKSWKSLISDSNF---------TKKNLRVSTTSHRLLFPKLTKG----QYIFNACT 596
Query: 88 TSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGLICLHGCFHGSGYKKHSFCFWN 147
S + + T ++ +Q+ + I GSC+G++CL + WN
Sbjct: 597 LSSLITTKGTATA------MQHPLNIRKFDKIRGSCHGILCLE-------LHQRFAILWN 737
Query: 148 P-ATRSKSKTLLYVPSYLNRVRLGFGYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVF 206
P + S L +P + N + FGYD+STD+YK F M V
Sbjct: 738 PFINKYASLPPLEIP-WSNTIYSCFGYDHSTDSYKVAAFIKWMPNS------EIYKTYVH 896
Query: 207 TLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVI 266
T+G + WR IQ FP L+S G ++S + +WL YK + ++
Sbjct: 897 TMGTTSWRMIQ-DFPCTPYLKS--------GKFVSWTFNWLA---YK---DKYSVSSLLV 1031
Query: 267 ISLDLETETYTQLQLP 282
+SL LE E+Y ++ P
Sbjct: 1032VSLHLENESYGEILQP 1079
>TC81783 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 1225
Score = 39.3 bits (90), Expect(3) = 2e-11
Identities = 15/40 (37%), Positives = 25/40 (62%)
Frame = +2
Query: 290 NICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
++ L DC+C Y+ + F +W MKE+G +ESWT+ +
Sbjct: 866 SVLMLRDCLCV-YEASDLFFNVWIMKEYGNQESWTKLYSV 982
Score = 37.7 bits (86), Expect(3) = 2e-11
Identities = 36/116 (31%), Positives = 47/116 (40%), Gaps = 6/116 (5%)
Frame = +3
Query: 122 SCNGLICLHGCFHGSGYKKHSFCF-WNPATRSKSKTLLYVPSYLNRVR-----LGFGYDN 175
SC+G+ C G +CF WNP+ R K K L P Y N + + FGYD+
Sbjct: 447 SCDGIFC--------GELNLGYCFVWNPSIR-KFKLL---PPYKNPLEGDPFSISFGYDH 590
Query: 176 STDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWD 231
D YK V + V V LG WR I+ P+ L S W+
Sbjct: 591 FIDNYKVVAISSKYE------------VFVNALGTDYWRRIR-EHPLLLLHPSTWN 719
Score = 35.8 bits (81), Expect = 0.027
Identities = 21/46 (45%), Positives = 29/46 (62%)
Frame = +2
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLS 52
LP +L+ EIL R V L+ L+ + K +N+LISDP F K H + S
Sbjct: 134 LPFDLLP-EILCRLPVKLLIQLRCLCKFFNSLISDPNFAKKHFQFS 268
Score = 28.1 bits (61), Expect(3) = 2e-11
Identities = 14/52 (26%), Positives = 26/52 (49%)
Frame = +1
Query: 237 GVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDN 288
G+++ +++W+ + I+SLDLE E+Y +L LP +N
Sbjct: 715 GIFVGGTVNWMASD---------IADLLFILSLDLEKESYQKLFLPDSENEN 843
>BF634925
Length = 679
Score = 61.6 bits (148), Expect = 5e-10
Identities = 31/63 (49%), Positives = 39/63 (61%), Gaps = 6/63 (9%)
Frame = +1
Query: 273 TETYTQLQLPKLPFDN------PNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQF 326
TE+YTQL +P FD ++C L D +CFS+D T F++WQMK GVE WTQ
Sbjct: 493 TESYTQL-MPPCGFDEMSYVKPASLCILKDSLCFSHDNWRTEFIVWQMKIIGVEXPWTQL 669
Query: 327 LKI 329
LKI
Sbjct: 670 LKI 678
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,951,430
Number of Sequences: 36976
Number of extensions: 244471
Number of successful extensions: 1500
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 1423
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1441
length of query: 362
length of database: 9,014,727
effective HSP length: 97
effective length of query: 265
effective length of database: 5,428,055
effective search space: 1438434575
effective search space used: 1438434575
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136954.5


