

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.9 - phase: 0
(567 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
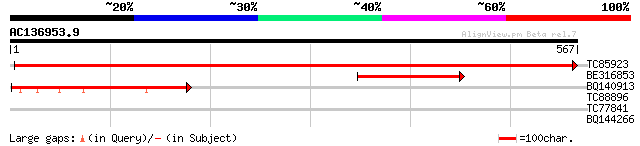
Score E
Sequences producing significant alignments: (bits) Value
TC85923 similar to SP|Q42876|AMP2_LYCES Aminopeptidase 2 chloro... 1085 0.0
BE316853 homologue to GP|15983519|gb AT4g30920/F6I18_170 {Arabid... 206 2e-53
BQ140913 similar to SP|Q42876|AMP2_ Aminopeptidase 2 chloroplas... 169 3e-42
TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown prot... 31 1.1
TC77841 weakly similar to GP|9294387|dbj|BAB02397.1 cytochrome P... 29 4.2
BQ144266 similar to GP|19887436|gb| Fe-S oxidoreductase family p... 29 5.6
>TC85923 similar to SP|Q42876|AMP2_LYCES Aminopeptidase 2 chloroplast
precursor (EC 3.4.11.1) (Leucine aminopeptidase) (LAP),
partial (89%)
Length = 1930
Score = 1085 bits (2805), Expect = 0.0
Identities = 562/563 (99%), Positives = 562/563 (99%)
Frame = +2
Query: 5 ASAIVIAFSKSSSSSLFLTSRIRFASFPKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKI 64
ASAIVIAFSKSSSSSLFLTSRIRFASFPKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKI
Sbjct: 2 ASAIVIAFSKSSSSSLFLTSRIRFASFPKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKI 181
Query: 65 SFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFS 124
SFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFS
Sbjct: 182 SFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFS 361
Query: 125 GKVGQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASS 184
GKVGQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASS
Sbjct: 362 GKVGQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASS 541
Query: 185 EGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAG 244
EGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAG
Sbjct: 542 EGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAG 721
Query: 245 DVSFGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLG 304
DVS GIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLG
Sbjct: 722 DVSSGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLG 901
Query: 305 VAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGG 364
VAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGG
Sbjct: 902 VAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGG 1081
Query: 365 SAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEG 424
SAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEG
Sbjct: 1082SAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEG 1261
Query: 425 RLTLADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGE 484
RLTLADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGE
Sbjct: 1262RLTLADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGE 1441
Query: 485 KLWRLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWN 544
KLWRLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWN
Sbjct: 1442KLWRLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWN 1621
Query: 545 DKKRSATGFGVSTLVEWVLKNSS 567
DKKRSATGFGVSTLVEWVLKNSS
Sbjct: 1622DKKRSATGFGVSTLVEWVLKNSS 1690
>BE316853 homologue to GP|15983519|gb AT4g30920/F6I18_170 {Arabidopsis
thaliana}, partial (18%)
Length = 324
Score = 206 bits (524), Expect = 2e-53
Identities = 100/107 (93%), Positives = 104/107 (96%)
Frame = +2
Query: 348 KTGPGCSIELMKFDMGGSAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVV 407
KTGPGC IELMKFDMGGSAAV GAAKA+GQIKPLGVEVHFIVAACENMISGTGMRPGD+V
Sbjct: 2 KTGPGCMIELMKFDMGGSAAVFGAAKAIGQIKPLGVEVHFIVAACENMISGTGMRPGDIV 181
Query: 408 TASNGKTIEVNNTDAEGRLTLADALVYACNQGVEKIIDLATLTGACV 454
TASNGKTIEVNNTDAEGRLTLADALVY CNQGV+KIIDLATLTGAC+
Sbjct: 182 TASNGKTIEVNNTDAEGRLTLADALVYTCNQGVDKIIDLATLTGACI 322
>BQ140913 similar to SP|Q42876|AMP2_ Aminopeptidase 2 chloroplast precursor
(EC 3.4.11.1) (Leucine aminopeptidase) (LAP), partial
(10%)
Length = 642
Score = 169 bits (428), Expect = 3e-42
Identities = 103/195 (52%), Positives = 134/195 (67%), Gaps = 15/195 (7%)
Frame = +2
Query: 2 AAIASAIV---IAFSKS-SSSSLFLTSRI--RFASFPKRSFHSTTKLMSQSRY----TTL 51
AA +S I+ + FS S SSS LF+ R+ R AS P R T ++ ++ TL
Sbjct: 20 AASSSTILTTSLVFSSSFSSSRLFIRRRLPLRSASSPLRRLCLTREVPVRNLIMPPRATL 199
Query: 52 GLTHPTNIEAPKISFSATDVD--VTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLK 109
G THP N E PKISF+A D+D V EWKGD+LAV VTEKD+TRD + +F+NSIL +D K
Sbjct: 200 GFTHPANTETPKISFAAKDIDTDVAEWKGDLLAVAVTEKDVTRDGELKFQNSILRTLDSK 379
Query: 110 LDGLLSEASSEEDFSGKVGQSTVVRI---KGLGSKRVGLIGLGQLPSTTALYKGLGEAVV 166
L GLL EASSEEDF+GK+GQST++RI GLGSKR+ L+GLG S+TA +K LGE+V
Sbjct: 380 LGGLLGEASSEEDFTGKLGQSTLLRITPAAGLGSKRLALLGLGPSASSTAAFKTLGESVA 559
Query: 167 AVAKSAQASNVAIVL 181
+ A A A++VA+V+
Sbjct: 560 SAANFAXAAHVAVVV 604
>TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 1097
Score = 31.2 bits (69), Expect = 1.1
Identities = 28/113 (24%), Positives = 47/113 (40%), Gaps = 1/113 (0%)
Frame = -3
Query: 16 SSSSLFLTSRIR-FASFPKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKISFSATDVDVT 74
+SSS+F + I F SFP F S+ M +R + + + FS DV +
Sbjct: 1032 ASSSVFFSKEINLFTSFPNDEFSSSVLFMHSTRSCSFCSHSALEFKISILEFSILDVRTS 853
Query: 75 EWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFSGKV 127
++ + +KS F++SI L +D S +S+ DF +
Sbjct: 852 --IS*CASLSMDSYFSLLASKSDFKSSIS*CASLSMDSYFSLLASKSDFKSSI 700
>TC77841 weakly similar to GP|9294387|dbj|BAB02397.1 cytochrome P450
{Arabidopsis thaliana}, partial (38%)
Length = 1299
Score = 29.3 bits (64), Expect = 4.2
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = +3
Query: 431 ALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGEKLWRL 489
A Y+CN+ + K +L + G C V + P + D + + +S GEKL++L
Sbjct: 552 AFSYSCNEMISKWKELLSSDGTCEVDVWPFLQNFTC--DVISRTAFGSSYAEGEKLFQL 722
>BQ144266 similar to GP|19887436|gb| Fe-S oxidoreductase family protein
{Methanopyrus kandleri AV19}, partial (2%)
Length = 1120
Score = 28.9 bits (63), Expect = 5.6
Identities = 24/90 (26%), Positives = 39/90 (42%), Gaps = 7/90 (7%)
Frame = +3
Query: 45 QSRYTTLGL-------THPTNIEAPKISFSATDVDVTEWKGDILAVGVTEKDLTRDAKSR 97
QS Y+T G P +I K+ F A VT W+G E + D KSR
Sbjct: 237 QSAYSTNGR**KR*NHPRPDHIRPGKVPFPARSRSVT-WRGQ-------ESRVKFDLKSR 392
Query: 98 FENSILNKIDLKLDGLLSEASSEEDFSGKV 127
+ ++ ++ L + + S S+ + GK+
Sbjct: 393 YIIAVTTRMHLCENNIFSGPSAADPLKGKI 482
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,933,743
Number of Sequences: 36976
Number of extensions: 166230
Number of successful extensions: 615
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 610
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 613
length of query: 567
length of database: 9,014,727
effective HSP length: 101
effective length of query: 466
effective length of database: 5,280,151
effective search space: 2460550366
effective search space used: 2460550366
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136953.9


