

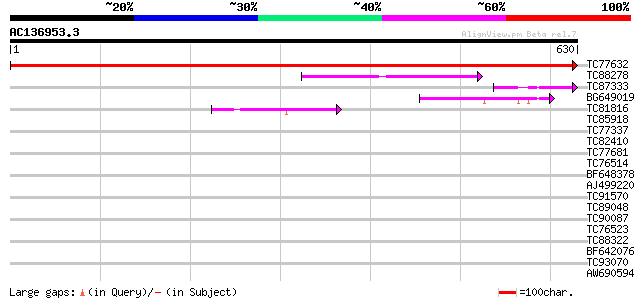
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.3 + phase: 0 /pseudo
(630 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77632 homologue to GP|18086492|gb|AAL57699.1 AT4g30600/F17I23_... 1212 0.0
TC88278 homologue to SP|P49972|SR52_LYCES Signal recognition par... 80 2e-15
TC87333 similar to SP|P49972|SR52_LYCES Signal recognition parti... 53 4e-07
BG649019 similar to PIR|T52612|T52 SRP receptor homolog FtsY pre... 51 1e-06
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 44 2e-04
TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreu... 40 0.003
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 38 0.013
TC82410 similar to GP|3850569|gb|AAC72109.1| ESTs gb|T21276 gb|... 37 0.023
TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone d... 37 0.030
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 37 0.030
BF648378 similar to GP|9294677|dbj| gene_id:F14O13.28~pir||T2263... 36 0.039
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 35 0.067
TC91570 weakly similar to GP|7208782|emb|CAB76913.1 hypothetical... 35 0.087
TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein... 35 0.11
TC90087 similar to PIR|T05841|T05841 spliceosome-associated prot... 35 0.11
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 34 0.15
TC88322 similar to GP|15810008|gb|AAL06931.1 At1g63980/F22C12_9 ... 34 0.19
BF642076 homologue to GP|23505128|em hypothetical protein {Plasm... 34 0.19
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 34 0.19
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 34 0.19
>TC77632 homologue to GP|18086492|gb|AAL57699.1 AT4g30600/F17I23_60
{Arabidopsis thaliana}, partial (75%)
Length = 2273
Score = 1212 bits (3137), Expect = 0.0
Identities = 621/631 (98%), Positives = 622/631 (98%), Gaps = 1/631 (0%)
Frame = +3
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK
Sbjct: 123 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 302
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEA 120
WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEA
Sbjct: 303 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEA 482
Query: 121 EARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRS 180
EARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRS
Sbjct: 483 EARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRS 662
Query: 181 AIVNNGNGYNLRS-NGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVT 239
AIVNN + GVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVT
Sbjct: 663 AIVNNR*PVII*GVTGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVT 842
Query: 240 KAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFS 299
KAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFS
Sbjct: 843 KAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFS 1022
Query: 300 SDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTK 359
SDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTK
Sbjct: 1023SDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTK 1202
Query: 360 NVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAA 419
NVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAA
Sbjct: 1203NVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAA 1382
Query: 420 KEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRL 479
KEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRL
Sbjct: 1383KEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRL 1562
Query: 480 QIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPD 539
QIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPD
Sbjct: 1563QIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPD 1742
Query: 540 LVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVY 599
LVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVY
Sbjct: 1743LVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVY 1922
Query: 600 ISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
ISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL
Sbjct: 1923ISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 2015
>TC88278 homologue to SP|P49972|SR52_LYCES Signal recognition particle 54
kDa protein 2 (SRP54). [Tomato] {Lycopersicon
esculentum}, partial (40%)
Length = 713
Score = 80.1 bits (196), Expect = 2e-15
Identities = 49/201 (24%), Positives = 95/201 (46%)
Frame = +3
Query: 325 GWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLAS 384
G + + ++ +++ L L + L+ +V ++ + ++ + LA+
Sbjct: 123 GSITRALKQMSNATIIDEKVLNECLNEITRALLQSDVQFKLVRDMQTNIKKIVNLDDLAA 302
Query: 385 FTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAK 444
+Q A+ + L IL P + ++ K VV+FVG+ G GK+T K
Sbjct: 303 GHNKRRIIQQAVFNELCNILDPGKPA-------FTPKKGKASVVMFVGLQGSGKTTTCTK 461
Query: 445 VAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEAS 504
A++ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E ++
Sbjct: 462 YAFYHQKKGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGVERFK 641
Query: 505 RNGSDVVLVDTAGRMQDNEPL 525
+ D+++VDT+GR + L
Sbjct: 642 KENCDLIIVDTSGRHKQEAAL 704
>TC87333 similar to SP|P49972|SR52_LYCES Signal recognition particle 54 kDa
protein 2 (SRP54). [Tomato] {Lycopersicon esculentum},
partial (58%)
Length = 1236
Score = 52.8 bits (125), Expect = 4e-07
Identities = 32/93 (34%), Positives = 49/93 (52%)
Frame = +1
Query: 538 PDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSM 597
PDLV+FV ++ +G A DQ F Q +A + +++TK D K G ALS
Sbjct: 34 PDLVIFVMDSSIGQAAFDQAQAFKQSVA----------VGAVIITKMDG-HAKGGGALSA 180
Query: 598 VYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
V + +PV+F+G G+ + + +VK V LL
Sbjct: 181 VAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLL 279
>BG649019 similar to PIR|T52612|T52 SRP receptor homolog FtsY precursor
chloroplast [validated] - Arabidopsis thaliana, partial
(39%)
Length = 788
Score = 51.2 bits (121), Expect = 1e-06
Identities = 51/172 (29%), Positives = 79/172 (45%), Gaps = 22/172 (12%)
Frame = +1
Query: 456 VMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPA-VVAKEAIQEASRNGSDVVLVD 514
+++AA DTFR+ A +QL A R I EK A V +A++ G D+VL D
Sbjct: 199 ILLAAGDTFRAAASDQLEIWAERTGCEIVVAESEKSKASAVLSQAVKRGKELGYDIVLCD 378
Query: 515 TAGRMQDNEPLM-------RALSKLIY-LNNPDLVLFVGEALVGNDAVDQLSKFNQKL-- 564
T+GR+ N LM +A++K++ N V F + LV + A + F ++
Sbjct: 379 TSGRLHTNYRLMEELISCKKAVAKVVAGAPNVSFVSFTLDILVLDTARAVAANFIMEILL 558
Query: 565 -----ADLSTSPTPRL------IDGILLTKFDTIDDKVGAALSMVYISGAPV 605
L+ P R + G++LTK D + G +S+V G PV
Sbjct: 559 VLDGTTGLNMLPQAREFNEVVGVTGLILTKLDG-SARGGCVVSVVDELGIPV 711
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 43.9 bits (102), Expect = 2e-04
Identities = 42/152 (27%), Positives = 62/152 (40%), Gaps = 8/152 (5%)
Frame = +3
Query: 225 RNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDR-KVDYL 283
+ K KK V + + KK +K K E K + + DGD KD+ K
Sbjct: 186 KEKKDKEKKDKTDVDEGKDKKDKEK------KKKEKKEENVKGEEEDGDEKKDKEKKKKE 347
Query: 284 AKEQGESMMDKDEIFSSDSEDEE-------DDDDDNAGKKSKPDAKKKGWFSSMFQSIAG 336
KE+G+ DKD +D+E DDD+ G K K + KK + G
Sbjct: 348 KKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEG 527
Query: 337 KANLEKSDLEPALKALKDRLMTKNVAEEIAEK 368
K ++ D+E K K++ K EE +K
Sbjct: 528 KVSVRDIDIEETAKEGKEKKKKKEDKEEKKKK 623
>TC85918 similar to GP|13562014|gb|AAK30610.1 fibroin 1 {Plectreurys tristis},
partial (8%)
Length = 4117
Score = 40.0 bits (92), Expect = 0.003
Identities = 29/120 (24%), Positives = 55/120 (45%)
Frame = -3
Query: 250 NRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDD 309
+++ DE E + + D D A K D + E+ E++ +K +++ D+ DD
Sbjct: 3587 SKIEDEVKAELAAEEIEKSD-DSSALKVSVEDIVKLEKDEAIAEKMNSLANEEGDKHDDK 3411
Query: 310 DDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKL 369
DN ++ P + +G ++ +E+SD ALK + ++ E IAEK+
Sbjct: 3410 PDNTQSQATPSSNTEG---DEVKTELAAGEIERSDDSSALKDFVEDIVNPEKDEAIAEKI 3240
Score = 39.7 bits (91), Expect = 0.004
Identities = 29/117 (24%), Positives = 56/117 (47%)
Frame = -3
Query: 265 TDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
T+ V++ D ++ +E+ E++ +K +++ D+ +D DNA ++ P +K
Sbjct: 3755 TETVELASDHASTTHEQFVKQEKDEAIAEKMNSLANEDGDKHNDKPDNAQGQATPSSK-- 3582
Query: 325 GWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKK 381
++ +EKSD ALK + ++ E IAEK+ + A+ EG K
Sbjct: 3581 --IEDEVKAELAAEEIEKSDDSSALKVSVEDIVKLEKDEAIAEKM--NSLANEEGDK 3423
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 37.7 bits (86), Expect = 0.013
Identities = 32/119 (26%), Positives = 56/119 (46%)
Frame = +3
Query: 206 ENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFT 265
E D +GAF + + GG G ++ K+ KK + ++ E + D
Sbjct: 444 EEDDDGDGAFGEGEDELSSEDGGGYGNNSN---NKSNSKKAPEGGAGGADENGEEE-DDE 611
Query: 266 DHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
D D D D D+D D +E+G +++ + D+E+EE+D+D+ A +P K+K
Sbjct: 612 DGDDQDEDDDEDEDDD--DEEEGGEEDEEEGVDEEDNEEEEEDEDEEA---LQPPKKRK 773
>TC82410 similar to GP|3850569|gb|AAC72109.1| ESTs gb|T21276 gb|T45403 and
gb|AA586113 come from this gene. {Arabidopsis thaliana},
partial (8%)
Length = 606
Score = 37.0 bits (84), Expect = 0.023
Identities = 32/140 (22%), Positives = 61/140 (42%), Gaps = 3/140 (2%)
Frame = +1
Query: 172 KKNSENDRSAIVNNGN---GYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKG 228
+K+ + S + N + G N S VV + + N V+ G D +++K ++
Sbjct: 28 EKDKSVEESRVFENKDDDGGVNKASKSVVDDAA-----NKEVSVGESDEKVIERKDGDEK 192
Query: 229 GNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQG 288
+ K +VV + +++ WD+ + +D +D KVD L +
Sbjct: 193 PSSKNESSVVESKHMENEEEEDLGWDDIEDLSGIDEKKETQSQSQSDSTSKVDLLKR--- 363
Query: 289 ESMMDKDEIFSSDSEDEEDD 308
S ++DE S D ED++D+
Sbjct: 364 LSTAEEDEDLSWDIEDDDDE 423
>TC77681 similar to GP|22597156|gb|AAN03465.1 nucleolar histone deacetylase
HD2-P39 {Glycine max}, partial (47%)
Length = 1233
Score = 36.6 bits (83), Expect = 0.030
Identities = 34/111 (30%), Positives = 50/111 (44%), Gaps = 7/111 (6%)
Frame = +2
Query: 241 AEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSS 300
+EPKK KN + K HV + D+D D + ++ S D+ E S
Sbjct: 482 SEPKKADAKNAAPAKNAATAK-----HVKVVDPKDEDSDDDDESDDEIGSSDDEMENADS 646
Query: 301 DSEDEEDDDDDNAGK---KSKPDAKKKGWFSSMFQSIAGK----ANLEKSD 344
DSEDE+D D+D+ + K KK+ S+ I+ K A EK+D
Sbjct: 647 DSEDEDDSDEDDEEETPVKKVDQGKKRPNESASKTPISSKKSKNATPEKTD 799
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 36.6 bits (83), Expect = 0.030
Identities = 42/180 (23%), Positives = 74/180 (40%), Gaps = 11/180 (6%)
Frame = +3
Query: 154 KNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGN-VSVNGKENDSVNN 212
KNGSA K D ++ ++S+ D ++ V + DS ++
Sbjct: 594 KNGSAPAK-KAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESS 770
Query: 213 GAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPV--------ETKLDF 264
D K +N KK + ++ + + + D KPV ++K D
Sbjct: 771 DEEDKKPAAKASKNVSAPTKKA---ASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDS 941
Query: 265 TDHVDIDGDA--DKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAK 322
+D D D D+ D+D+K AK++ + +DKD S SE+E +D+ +K D +
Sbjct: 942 SDSDDEDDDSSSDEDKK-PVAAKKEDKMNVDKDGSDSDQSEEESEDEPSKTPQKKIKDVE 1118
Score = 35.8 bits (81), Expect = 0.051
Identities = 46/183 (25%), Positives = 78/183 (42%), Gaps = 12/183 (6%)
Frame = +3
Query: 154 KNGSAGGGLKNDGDGKNGKKNSENDR---SAIVNNGNGYNLRSNGVVGNVSVNGKENDSV 210
KNGSA + + ++ +++S+ D+ + V + NG S S ++S
Sbjct: 489 KNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNG----SAPAKKAASDEEDTDESS 656
Query: 211 NNGAFDVNRLQKKVRNKGGN--GKKTD-------AVVTKAEPKKVVKKNRVWDEKPVETK 261
+ D K V +K G+ KK D + + E KK K P +
Sbjct: 657 DEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKA 836
Query: 262 LDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDA 321
+D + D ++D+D ++K + K + SSDS+DE DDD ++ + KP A
Sbjct: 837 ASSSDE-ESDEESDEDEDAKPVSKPAAVAKKSKKD--SSDSDDE--DDDSSSDEDKKPVA 1001
Query: 322 KKK 324
KK
Sbjct: 1002AKK 1010
>BF648378 similar to GP|9294677|dbj| gene_id:F14O13.28~pir||T22638~similar to
unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 651
Score = 36.2 bits (82), Expect = 0.039
Identities = 30/128 (23%), Positives = 57/128 (44%), Gaps = 1/128 (0%)
Frame = +2
Query: 191 LRSNGVVGNVS-VNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKK 249
L+ GV+G+ S E++S ++ DVN K + K+ + K + K V K
Sbjct: 272 LKKKGVIGSPSHTEDDESESESSSDDDVNNFNTK------SDKEFFDALIKVKKKDPVLK 433
Query: 250 NRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDD 309
+ D K E+ D D + DK++K +L + ++++ F + ++ +
Sbjct: 434 QK--DVKLFESDHSSEDESDDEKSKDKEKKSMFLKDVVAKHLIEEGPDFGDEEDETNEIG 607
Query: 310 DDNAGKKS 317
N GKK+
Sbjct: 608 ISNGGKKT 631
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (12%)
Length = 518
Score = 35.4 bits (80), Expect = 0.067
Identities = 21/91 (23%), Positives = 48/91 (52%)
Frame = -3
Query: 283 LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEK 342
+ +E+ ES MD+ E + D +++ED+ +++ + ++ + ++ MFQ A + ++
Sbjct: 492 MQREEDESAMDRGEYYDEDEDEDEDEYEEDGEEDNQTEEQRMEEGRRMFQIFAARMFEQR 313
Query: 343 SDLEPALKALKDRLMTKNVAEEIAEKLCESV 373
L A +++ VA+E +KL E +
Sbjct: 312 -----VLNAYREK-----VAQERQQKLLEEL 250
>TC91570 weakly similar to GP|7208782|emb|CAB76913.1 hypothetical protein
{Cicer arietinum}, partial (17%)
Length = 1446
Score = 35.0 bits (79), Expect = 0.087
Identities = 45/214 (21%), Positives = 82/214 (38%), Gaps = 10/214 (4%)
Frame = +2
Query: 110 DEIFKQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKG--DGSDGKKNGSAGGGLKNDGD 167
D Q K E + K SN GG ++++V K + + K S G K++
Sbjct: 77 DXCLSQRKYLEENNDQPKKNSN----GGAKEEKVLGKKTLEEKNDKPKKSLNGSAKDEKS 244
Query: 168 GKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNK 227
+ ND+ NG+ +S G N K S+N G + L +K + +
Sbjct: 245 LEQKTLEENNDKPKKSLNGSAKEEKSLGQKTPEENNDKPKKSLNGGVKEEKDLGQKTQEE 424
Query: 228 GGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQ 287
+ K E K + +K + ++KP ++ + G A +++ + A+E+
Sbjct: 425 NNDKPKKSLNGGVKEEKDLGQKTQENNDKPKKS---------LSGGAKEEKGLGQKAQEE 577
Query: 288 GESMMDKDEIFSSDSE--------DEEDDDDDNA 313
K S+ E ++E +D NA
Sbjct: 578 NNGKPKKSLNGSAKEEKGLRQKALEQEGHNDPNA 679
>TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein T8P19.220
- Arabidopsis thaliana, partial (35%)
Length = 1329
Score = 34.7 bits (78), Expect = 0.11
Identities = 21/83 (25%), Positives = 41/83 (49%)
Frame = +2
Query: 242 EPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSD 301
+PK+V +N DE + +++ T+ ++D+ +++D G DKD+
Sbjct: 332 KPKQV--ENEAKDEPKPKEEVEVTEQDAEVAESDEKKEID------GHEDSDKDDEDEGG 487
Query: 302 SEDEEDDDDDNAGKKSKPDAKKK 324
ED+ DDD++ + K KK+
Sbjct: 488 DEDDADDDEEEDAGEEKKGVKKE 556
>TC90087 similar to PIR|T05841|T05841 spliceosome-associated protein homolog
F17L22.120 - Arabidopsis thaliana, partial (12%)
Length = 733
Score = 34.7 bits (78), Expect = 0.11
Identities = 37/158 (23%), Positives = 66/158 (41%), Gaps = 24/158 (15%)
Frame = +2
Query: 191 LRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVR---------NKGGNGKKTDA----- 236
+ NGVV N + K + D R ++K + N + TDA
Sbjct: 113 VHQNGVVSNGDLGSKTATVKKSRESDRRRRRRKQKKINKASKEQNANASEDDTDAKENTE 292
Query: 237 -----VVTKAEPKKVVKKNRVWDE-----KPVETKLDFTDHVDIDGDADKDRKVDYLAKE 286
VV + E + V +K +++ K + K F++ +D + + KD E
Sbjct: 293 QQQQQVVEQVEIEYVPEKAELYEGLDEEFKKIFEKFSFSEVIDSEDNDKKD--------E 448
Query: 287 QGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
E+ + K + +DS+ EE++DD+ +K + KKK
Sbjct: 449 SAENAITKKK---ADSDSEEEEDDNEQKEKGVSNKKKK 553
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 34.3 bits (77), Expect = 0.15
Identities = 38/144 (26%), Positives = 65/144 (44%), Gaps = 8/144 (5%)
Frame = +3
Query: 246 VVKKNRVWDEKPV------ETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFS 299
++KK R +KP+ + K + + D ++D+D ++K + K + S
Sbjct: 90 LMKKIRNLQQKPLKM*VHPQRKPASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKD--S 263
Query: 300 SDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEK--SDLEPALKALKDRLM 357
SDS+DE+DD + KK K+ S K N++K SD + + + +D
Sbjct: 264 SDSDDEDDDSSSDEDKKPVAAKKEVSESESDSSDDEDKMNVDKDGSDSDESEEESEDE-P 440
Query: 358 TKNVAEEIAEKLCESVAASLEGKK 381
+K ++I K E V A GKK
Sbjct: 441 SKTPQKKI--KDVEMVDAGKSGKK 506
>TC88322 similar to GP|15810008|gb|AAL06931.1 At1g63980/F22C12_9
{Arabidopsis thaliana}, partial (54%)
Length = 1373
Score = 33.9 bits (76), Expect = 0.19
Identities = 36/174 (20%), Positives = 66/174 (37%), Gaps = 12/174 (6%)
Frame = +1
Query: 186 GNGYNLRSNGVVGNVSVNGKEN------DSVNNGAFDVNRLQ---KKVRNKGGNGKKTDA 236
G G G+ G+V V K++ + N AFD + K+++ + T++
Sbjct: 172 GEGLGKEKQGIKGHVRVTNKQDTTGIGLEKPNQWAFDTTQFDNILKRLKVQAPVSNDTES 351
Query: 237 VVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDG---DADKDRKVDYLAKEQGESMMD 293
K E + VV PV+ K+ G ++ + V + + + E ++
Sbjct: 352 NTVKVETEAVV---------PVDPKVAVPRATRPQGRYKKRERGKLVSHYSSKDLEGILV 504
Query: 294 KDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEP 347
K S + D DD+ D + K GW +F + K + S + P
Sbjct: 505 KKSDISEEGTDNADDELDLSKSPLHLKLLKVGWRELLFLNKIKKTSTI*SKINP 666
>BF642076 homologue to GP|23505128|em hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 503
Score = 33.9 bits (76), Expect = 0.19
Identities = 15/40 (37%), Positives = 27/40 (67%)
Frame = +3
Query: 273 DADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDN 312
+++ RK+D KE+G ++K E+ +D ED+ DDDD++
Sbjct: 123 ESEGARKMDDGKKEEGLRDLEKGEVGKNDVEDDLDDDDED 242
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 33.9 bits (76), Expect = 0.19
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 9/89 (10%)
Frame = +1
Query: 245 KVVKKNRV---------WDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKD 295
K++K NR +D ++DF D D D D D+D D D
Sbjct: 166 KILKTNRKSTYGKFLSPYDSDDEIEEMDFEDDEDEDEDEDED---------------DDD 300
Query: 296 EIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
+ D +D+EDDDDD + + +AK K
Sbjct: 301 D----DDDDDEDDDDDGFAEPTDLNAKDK 375
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 33.9 bits (76), Expect = 0.19
Identities = 20/68 (29%), Positives = 38/68 (55%)
Frame = +3
Query: 244 KKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSE 303
+KV + N DE VE + D D + D + D++ + + +E+ E +++E + E
Sbjct: 36 RKVEEHNEEEDEGEVEEEEDEHDEEEED-EHDEEEEEEEEEEEEDEHDDEEEEEEEEEEE 212
Query: 304 DEEDDDDD 311
+EEDDD++
Sbjct: 213 EEEDDDEE 236
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,158,882
Number of Sequences: 36976
Number of extensions: 178824
Number of successful extensions: 1705
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 1208
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1421
length of query: 630
length of database: 9,014,727
effective HSP length: 102
effective length of query: 528
effective length of database: 5,243,175
effective search space: 2768396400
effective search space used: 2768396400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136953.3


