

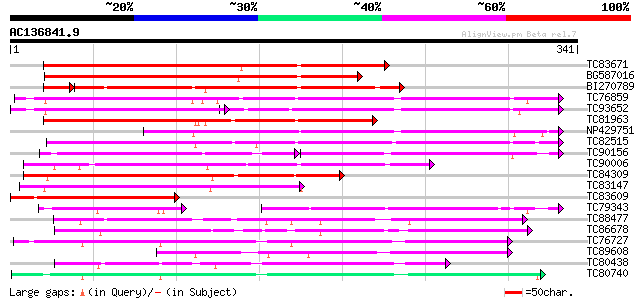
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.9 + phase: 0
(341 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83671 similar to PIR|T09416|T09416 coil protein PO22 microspo... 302 1e-82
BG587016 similar to PIR|T09416|T09 coil protein PO22 microspore... 283 5e-77
BI270789 similar to PIR|T09416|T09 coil protein PO22 microspore... 221 4e-64
TC76859 Enod8.1 [Medicago truncatula]; early nodule-specific pro... 216 1e-56
TC93652 Enod8.2 [Medicago truncatula] 126 5e-54
TC81963 Enod8-like protein [Medicago truncatula] 178 2e-45
NP429751 NP429751|AF463407.1|AAL68830.1 Enod8.3 [Medicago trunca... 176 2e-44
TC82515 similar to GP|3688284|emb|CAA09694.1 lanatoside 15'-O-ac... 176 2e-44
TC90156 similar to PIR|B86227|B86227 hypothetical protein [impor... 102 3e-43
TC90006 similar to PIR|A96590|A96590 hypothetical protein T22H22... 169 1e-42
TC84309 similar to GP|9294302|dbj|BAB02204.1 nodulin-like protei... 153 8e-38
TC83147 similar to PIR|T48618|T48618 early nodule-specific prote... 152 2e-37
TC83609 similar to GP|5295941|dbj|BAA81842.1 ESTs AU075322(C1110... 100 1e-21
TC79343 weakly similar to PIR|E86411|E86411 protein F1K23.18 [im... 86 3e-17
TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase p... 85 5e-17
TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20... 83 1e-16
TC76727 similar to GP|10638955|emb|CAB81548. putative proline-ri... 83 1e-16
TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/... 80 9e-16
TC80438 similar to PIR|E96579|E96579 hypothetical protein T18A20... 75 4e-14
TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif lipase/h... 73 2e-13
>TC83671 similar to PIR|T09416|T09416 coil protein PO22
microspore/pollen-specific - alfalfa, partial (60%)
Length = 671
Score = 302 bits (774), Expect = 1e-82
Identities = 151/212 (71%), Positives = 177/212 (83%), Gaps = 4/212 (1%)
Frame = +3
Query: 21 SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELE 80
S KCVYPAIYNFGDSNSDTG YAT +V+ PNGI+FFG+ISGR DGRLI+D I+EEL+
Sbjct: 42 SDKCVYPAIYNFGDSNSDTGTIYATYTSVQPPNGITFFGNISGRASDGRLIIDSITEELK 221
Query: 81 LPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQ-- 138
LPYLS+YLNSVGSNYRHGANFAV+ A IRP L LG QVSQFILFKSHTKILF+Q
Sbjct: 222 LPYLSAYLNSVGSNYRHGANFAVSGASIRPRGYHLFNLGLQVSQFILFKSHTKILFNQLS 401
Query: 139 --RTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQA 196
RTEP L+SG+PR EDFSKA+YTIDIGQND+ +G Q +SEE+V+RSIP+ILS F+Q+
Sbjct: 402 NNRTEPSLKSGLPRPEDFSKALYTIDIGQNDLAHGFQ--YTSEEQVQRSIPEILSNFSQS 575
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPH 228
V++LYNE ARVFWIHNTGPI C+P+ Y+ Y H
Sbjct: 576 VKQLYNEGARVFWIHNTGPIGCLPFNYYTYKH 671
>BG587016 similar to PIR|T09416|T09 coil protein PO22
microspore/pollen-specific - alfalfa, partial (55%)
Length = 696
Score = 283 bits (725), Expect = 5e-77
Identities = 144/195 (73%), Positives = 161/195 (81%), Gaps = 4/195 (2%)
Frame = +3
Query: 22 KKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELEL 81
KKC YPAIYNFGDSNSDTGA A AV PNGIS+FGS +GR DGRLI+DFISEEL+L
Sbjct: 114 KKCEYPAIYNFGDSNSDTGAANAIYTAVTPPNGISYFGSTTGRASDGRLIIDFISEELKL 293
Query: 82 PYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILF----D 137
PYLS+YLNS+GSNYRHGANFAV A IRP +LG QVSQFILFKSHTKILF D
Sbjct: 294 PYLSAYLNSIGSNYRHGANFAVGGASIRPGGYSPIFLGLQVSQFILFKSHTKILFNQLSD 473
Query: 138 QRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAV 197
RTE P +SG+PR E+FSKA+YTIDIGQND+ GLQ N+SEE+V+RSIPDILSQF+QAV
Sbjct: 474 NRTESPFKSGLPRNEEFSKALYTIDIGQNDLAIGLQ--NTSEEQVKRSIPDILSQFSQAV 647
Query: 198 QKLYNEEARVFWIHN 212
Q+LYNE ARVFWIHN
Sbjct: 648 QQLYNEGARVFWIHN 692
Score = 31.6 bits (70), Expect = 0.46
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = +1
Query: 1 MNTMTLIYILCFFNLCVACPSKKCVYPAIYN 31
MN+M LI++L FFNLCV + + +YN
Sbjct: 19 MNSMRLIHVLWFFNLCVTFTFIQVLSENVYN 111
>BI270789 similar to PIR|T09416|T09 coil protein PO22
microspore/pollen-specific - alfalfa, partial (62%)
Length = 657
Score = 221 bits (564), Expect(2) = 4e-64
Identities = 117/200 (58%), Positives = 148/200 (73%), Gaps = 2/200 (1%)
Frame = +1
Query: 40 GAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGA 99
G YAT + PNGISF G+ISGR DGRLI+D+I+EEL++PYLS+YLNSVGSNYR+GA
Sbjct: 76 GTAYATFLCNQPPNGISF-GNISGRASDGRLIIDYITEELKVPYLSAYLNSVGSNYRYGA 252
Query: 100 NFAVASAPIRPIIAGLT--YLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKA 157
NFA A IRP +G + +LG QV QFI FKSHT+ILF+ TEP L+SG+PR EDF A
Sbjct: 253 NFAAGGASIRP-GSGFSPFHLGLQVDQFIQFKSHTRILFNNGTEPSLKSGLPRPEDFCTA 429
Query: 158 IYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIE 217
+YTIDIG ND+ G ++SEE+V+ S P+IL F++AV++LYN ARVFWIHN GP+
Sbjct: 430 LYTIDIGLNDLASGFL--HASEEQVQMSFPEILGHFSKAVKQLYNVXARVFWIHNVGPVG 603
Query: 218 CIPYYYFFYPHKNEKGNLDA 237
C P P+ N+KG L+A
Sbjct: 604 CCP--SIITPNXNKKGILNA 657
Score = 41.2 bits (95), Expect(2) = 4e-64
Identities = 17/19 (89%), Positives = 18/19 (94%)
Frame = +3
Query: 21 SKKCVYPAIYNFGDSNSDT 39
S +CVYPAIYNFGDSNSDT
Sbjct: 18 SHECVYPAIYNFGDSNSDT 74
>TC76859 Enod8.1 [Medicago truncatula]; early nodule-specific protein
Length = 1350
Score = 216 bits (549), Expect = 1e-56
Identities = 135/344 (39%), Positives = 193/344 (55%), Gaps = 14/344 (4%)
Frame = +2
Query: 4 MTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGS 60
+TLI ++ LC+ P +K C +PAI++FG SN DTG A A P G ++F
Sbjct: 83 VTLIVLV----LCITPPIFATKNCDFPAIFSFGASNVDTGGLAAAFRAPPSPYGETYFHR 250
Query: 61 ISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI---RPIIAG--L 115
+GR DGR+ILDFI+ LPYLS YLNS+GSN+ HGANFA + I + I+ L
Sbjct: 251 STGRFSDGRIILDFIARSFRLPYLSPYLNSLGSNFTHGANFASGGSTINIPKSILPNGKL 430
Query: 116 TYLGFQVS--QFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQ 173
+ Q+ QF F S TK++ DQ + +P+ + FSKA+Y DIGQND+ G
Sbjct: 431 SPFSLQIQYIQFKEFISKTKLIRDQ--GGVFATLIPKEDYFSKALYIFDIGQNDLTIGF- 601
Query: 174 KPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKG 233
N + ++V ++PDI++ + + ++ +YN AR FWIH TGP C P +P +
Sbjct: 602 FGNKTIQQVNATVPDIVNNYIENIKNIYNLGARSFWIHGTGPKGCAPVILANFPSAIK-- 775
Query: 234 NLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFV 293
D+ GC K +NE++Q +N +LK+ + +LR A TYVD+YT KY+L +N GF
Sbjct: 776 --DSYGCAKQYNEVSQYFNFKLKEALAELRSNLSSAAITYVDIYTPKYSLFTNPEKYGFE 949
Query: 294 NPLEFCCGSYQGNEIHY---CGKKSIKNGT-VYGIACDDSSTYI 333
P CCG G E + CG NGT + +C + ST I
Sbjct: 950 LPFVACCG--YGGEYNIGVGCGASININGTKIVAGSCKNPSTRI 1075
>TC93652 Enod8.2 [Medicago truncatula]
Length = 1195
Score = 126 bits (317), Expect(2) = 5e-54
Identities = 80/210 (38%), Positives = 117/210 (55%), Gaps = 3/210 (1%)
Frame = +2
Query: 127 LFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSI 186
+F S T ++ DQ + +P+ + FSKA+YT DIGQND+ G N + ++V ++
Sbjct: 464 IFISKTNLIRDQGGV--FATLIPKEDYFSKALYTFDIGQNDL-IGGYFGNKTIKQVNATV 634
Query: 187 PDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNE 246
PDI++ F ++ +YN AR FWIH+T P C P +P + D+ GC K +NE
Sbjct: 635 PDIVNNFIVNIKNIYNLGARSFWIHSTVPSGCTPTILANFPSAIK----DSYGCAKQYNE 802
Query: 247 LAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQG- 305
++Q +N +LK + QLR PLA TYVD+Y+ Y+L N + GF P CCG Y G
Sbjct: 803 VSQYFNLKLKKALAQLRVDLPLAAITYVDIYSPNYSLFQNPKKYGFELPHVACCG-YGGK 979
Query: 306 -NEIHYCGKKSIKNGT-VYGIACDDSSTYI 333
N CG+ NGT + +C + ST I
Sbjct: 980 YNIRVGCGETLNINGTKIEAGSCKNPSTRI 1069
Score = 102 bits (254), Expect(2) = 5e-54
Identities = 57/136 (41%), Positives = 80/136 (57%), Gaps = 4/136 (2%)
Frame = +3
Query: 1 MNTMTLIYILCFFNLCVACP----SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGIS 56
M+ ++LI ++ LC+ P ++ C +PAI++FG SN DTG A A P G +
Sbjct: 60 MSLVSLIVLI----LCIITPPIFATRNCDFPAIFSFGASNVDTGGLAAAFQAPPSPYGET 227
Query: 57 FFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLT 116
+F +GR DGR+ILDFI++ LPYLS YLNS+GSN+ HGANFA + I T
Sbjct: 228 YFHRSTGRFSDGRIILDFIAQSFGLPYLSPYLNSLGSNFTHGANFATGGSTINSP**YYT 407
Query: 117 YLGFQVSQFILFKSHT 132
+G+ V FK H+
Sbjct: 408 VMGYLVHFLFKFKVHS 455
>TC81963 Enod8-like protein [Medicago truncatula]
Length = 783
Score = 178 bits (452), Expect = 2e-45
Identities = 99/208 (47%), Positives = 138/208 (65%), Gaps = 7/208 (3%)
Frame = +3
Query: 21 SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELE 80
+ +C + AI+NFGDSNSDTG A+ A + P G ++F +GR DGRLI+DFI++
Sbjct: 168 NSECNFRAIFNFGDSNSDTGGLAASFVAPKPPYGETYFHRPNGRFSDGRLIVDFIAQSFG 347
Query: 81 LPYLSSYLNSVGSNYRHGANFAVASAPIRP---II--AGLT--YLGFQVSQFILFKSHTK 133
LPYLS+YL+S+G+N+ HGANFA S+ IRP II G + YL Q +QF FK T+
Sbjct: 348 LPYLSAYLDSLGTNFSHGANFATTSSTIRPPPSIIPQGGFSPFYLDVQYTQFRDFKPRTQ 527
Query: 134 ILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQF 193
F ++ S +P+ E FSKA+YT DIGQND+G G N + ++V S+P+I++ F
Sbjct: 528 --FIRQQGGLFASLMPKEEYFSKALYTFDIGQNDLGAGF-FGNMTIQQVNASVPEIINSF 698
Query: 194 TQAVQKLYNEEARVFWIHNTGPIECIPY 221
++ V+ +YN R FWIHNTGPI C+PY
Sbjct: 699 SKNVKDIYNLGGRSFWIHNTGPIGCLPY 782
>NP429751 NP429751|AF463407.1|AAL68830.1 Enod8.3 [Medicago truncatula]
Length = 902
Score = 176 bits (445), Expect = 2e-44
Identities = 109/263 (41%), Positives = 149/263 (56%), Gaps = 10/263 (3%)
Frame = +3
Query: 81 LPYLSSYLNSVGSNYRHGANFAVASAPIR---PIIAGLTYLGFQVS-QFILFKSHT-KIL 135
LPYLS YLNS+GSN+ HGANFA A + I+ II + F + Q I FK K
Sbjct: 15 LPYLSPYLNSLGSNFTHGANFATAGSTIKIPNSIIPNGMFSPFSLQIQSIQFKDFIPKAK 194
Query: 136 FDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQ 195
F + + +P+ + +SKA+YT DIGQND+ G N + ++V ++PDI+ F
Sbjct: 195 FIRDQGGVFATLIPKEDYYSKALYTFDIGQNDLTAGFFG-NKTIQQVNTTVPDIVKSFID 371
Query: 196 AVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQL 255
++ +YN AR FWIHNTGPI C+P +P + D GC K +NE++Q +N +L
Sbjct: 372 NIKNIYNLGARSFWIHNTGPIGCVPLILANFPSAIK----DRYGCAKQYNEVSQYFNLKL 539
Query: 256 KDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGS---YQGNEIHYCG 312
K+ + QLR+ PLA TYVD+Y+ KY+L N + GF PL CCG+ Y N CG
Sbjct: 540 KEALAQLRKDLPLAAITYVDIYSPKYSLFQNPKKYGFELPLVACCGNGGKYNYNIRAGCG 719
Query: 313 KKSIKNG--TVYGIACDDSSTYI 333
NG TV G +C ST I
Sbjct: 720 ATININGTNTVVG-SCKKPSTRI 785
>TC82515 similar to GP|3688284|emb|CAA09694.1 lanatoside
15'-O-acetylesterase {Digitalis lanata}, partial (76%)
Length = 1230
Score = 176 bits (445), Expect = 2e-44
Identities = 108/324 (33%), Positives = 167/324 (51%), Gaps = 13/324 (4%)
Frame = +1
Query: 23 KCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELP 82
KC + I+NFGDSNSDTG Y+ A P G+++F + GR DGRLI+DF++E L LP
Sbjct: 130 KCDFQGIFNFGDSNSDTGGFYSAFPAQPIPYGMTYFKTPVGRSSDGRLIVDFLAEALGLP 309
Query: 83 YLSSYLNSVGSNYRHGANFAV-ASAPIRP----IIAGLTYLGFQVSQFILFKSHTKILFD 137
YLS YL S+GS+Y HGANFA AS + P ++GL+ Q+ + + K+ D
Sbjct: 310 YLSPYLQSIGSDYTHGANFATSASTVLLPTTSLFVSGLSPFALQIQLRQMQQFRAKV-HD 486
Query: 138 QRTEPPLRSG-------VPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDIL 190
PL+ +P + F K+IY IGQND + + ++ +P I+
Sbjct: 487 FHKRDPLKPSTCASKIKIPSPDIFGKSIYMFYIGQNDFTSKI-AASGGINGLKNYLPQII 663
Query: 191 SQFTQAVQKLYNEE-ARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQ 249
Q A+++LY + R F + N GP+ C P Y PH + +L+ +GC+ +N
Sbjct: 664 YQIASAIKELYYAQGGRTFMVLNLGPVGCYPGYLVELPHTS--SDLNEHGCIITYNNAVD 837
Query: 250 EYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIH 309
+YN+ LK+ + Q R+ A YVD + L + + G + + CCG + G + +
Sbjct: 838 DYNKLLKETLTQTRKSLSDASLIYVDTNSALMELFRHPTSYGLKHSTKACCG-HGGGDYN 1014
Query: 310 YCGKKSIKNGTVYGIACDDSSTYI 333
+ K G + AC+D Y+
Sbjct: 1015FDPKALC--GNMLASACEDPQNYV 1080
>TC90156 similar to PIR|B86227|B86227 hypothetical protein [imported] -
Arabidopsis thaliana, partial (84%)
Length = 1421
Score = 102 bits (255), Expect(2) = 3e-43
Identities = 57/162 (35%), Positives = 82/162 (50%), Gaps = 4/162 (2%)
Frame = +1
Query: 176 NSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNL 235
N S +V + IP ++++ AV+ LYNE R FW+HNTGP C+P + K +L
Sbjct: 571 NLSYVQVIKRIPTVITEIENAVKSLYNEGGRKFWVHNTGPFGCLPKLIAL----SXKKDL 738
Query: 236 DANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNP 295
D+ GC+ +N A+ +N L +LR A YVD+Y +K LI+NA GF NP
Sbjct: 739 DSFGCLSSYNSAARLFNEALYHSSQKLRTELKDATLVYVDIYAIKNDLITNATKYGFTNP 918
Query: 296 LEFCCG----SYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
L CCG Y + CG+ + CD+ S Y+
Sbjct: 919 LMVCCGFGGPPYNFDARVTCGQPGYQ-------VCDEGSRYV 1023
Score = 90.5 bits (223), Expect(2) = 3e-43
Identities = 60/158 (37%), Positives = 85/158 (52%), Gaps = 2/158 (1%)
Frame = +3
Query: 19 CPSKKCVYPAIYNFGDSNSDTGAGYATMA-AVEHPNGISFFGSISGRCCDGRLILDFISE 77
C SK V ++ FGDSNSDTG + + V PNG +FF +GR DGRL++DF+ +
Sbjct: 123 CSSKPAV---VFVFGDSNSDTGGLVSGLGFPVNLPNGRTFFHRSTGRLSDGRLVIDFLCQ 293
Query: 78 ELELPYLSSYLNSV-GSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILF 136
L +L+ YL+S+ GS + +GANFAV + P + L QV QF FK+ + L
Sbjct: 294 SLNTRFLTPYLDSMSGSTFTNGANFAVVGSSTLPKYLPFS-LNIQVMQFQHFKARSLQLA 470
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQK 174
+ + + F A+Y IDIGQND+ K
Sbjct: 471 TSGAKNMIND-----QGFRDALYLIDIGQNDLADSFTK 569
>TC90006 similar to PIR|A96590|A96590 hypothetical protein T22H22.20
[imported] - Arabidopsis thaliana, partial (63%)
Length = 858
Score = 169 bits (428), Expect = 1e-42
Identities = 101/258 (39%), Positives = 144/258 (55%), Gaps = 11/258 (4%)
Frame = +3
Query: 9 ILCFFNLCVACPSKKCV---YPAIYNFGDSNSDTG----AGYATMAAVEHPNGISFFGSI 61
I+C P K + +PA++NFGDSNSDTG AG+ ++ PNG ++F
Sbjct: 57 IICIITTIFLLPCAKSIHLDFPAVFNFGDSNSDTGTLVTAGFESLYP---PNGHTYFHLP 227
Query: 62 SGRCCDGRLILDFISEELELPYLSSYLNSVG-SNYRHGANFAVASAPIRPIIAGLTY--- 117
SGR DGRLI+DF+ + L+LP+L++YL+S+G N+R G NFA A + I P A
Sbjct: 228 SGRYSDGRLIIDFLMDALDLPFLNAYLDSLGLPNFRKGCNFAAAGSTILPATASSICPFS 407
Query: 118 LGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNS 177
G QVSQF+ FK+ L + VP + F K +Y DIGQND+ +
Sbjct: 408 FGIQVSQFLKFKARALELLSGKGRK-FDKYVPSEDIFEKGLYMFDIGQNDLAGAFY--SK 578
Query: 178 SEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDA 237
+ ++V SIP IL +F +++LY+E AR FWIHNTGP+ C+ + + LD
Sbjct: 579 TLDQVLASIPTILLEFESGIKRLYDEGARYFWIHNTGPLGCLAQNVAKF--GTDPSKLDE 752
Query: 238 NGCVKPHNELAQEYNRQL 255
GCV HN+ + +N QL
Sbjct: 753 LGCVSGHNQAVKTFNLQL 806
>TC84309 similar to GP|9294302|dbj|BAB02204.1 nodulin-like protein protein
{Arabidopsis thaliana}, partial (47%)
Length = 672
Score = 153 bits (387), Expect = 8e-38
Identities = 86/205 (41%), Positives = 127/205 (61%), Gaps = 12/205 (5%)
Frame = +2
Query: 9 ILCFFNLCVACPS-----KKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISG 63
++ F + V PS + C +PAI+NFGDSNSDTG A PNGI+FF + +G
Sbjct: 59 VISTFLVVVMVPSPVIGARNCSFPAIFNFGDSNSDTGGLSAAFGQAPPPNGITFFQTPAG 238
Query: 64 RCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGF--- 120
R DGRLI+DF+++ L LPYLS+YL+SVGSN+ +GANFA A + IRP + G+
Sbjct: 239 RFSDGRLIIDFLAQNLSLPYLSAYLDSVGSNFSNGANFATAGSTIRPQNTTKSQSGYSPI 418
Query: 121 ----QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPN 176
Q+ Q+ FK+ + ++ ++ +P+ E FS+A+YT DIGQND+ G K N
Sbjct: 419 SLDVQLIQYSDFKARSILV--RKKGGVFMKLLPKEEYFSEALYTFDIGQNDLTAG-YKLN 589
Query: 177 SSEEEVRRSIPDILSQFTQAVQKLY 201
+ E+V+ IPD+L QF+ ++ +Y
Sbjct: 590 MTTEQVKAYIPDVLGQFSDVIRSVY 664
>TC83147 similar to PIR|T48618|T48618 early nodule-specific protein-like -
Arabidopsis thaliana, partial (43%)
Length = 653
Score = 152 bits (384), Expect = 2e-37
Identities = 86/188 (45%), Positives = 113/188 (59%), Gaps = 17/188 (9%)
Frame = +1
Query: 7 IYILCFFNL-CVAC-----PSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGS 60
++++ FF L CV C S C +PAIYNFGDSNSDTG A + P G FF
Sbjct: 13 VFVVFFFFLSCVKCVELKDSSPSCSFPAIYNFGDSNSDTGGISAAFVPIPSPYGQGFFHK 192
Query: 61 ISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLG- 119
GR DGR+ILD+I+++L+ PYLS+YLNS+G+NYRHGANFA + IR + G
Sbjct: 193 PFGRDSDGRVILDYIADKLKWPYLSAYLNSLGTNYRHGANFATGGSTIRKQNETIFQYGI 372
Query: 120 ------FQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQ 173
Q+ QF FK+ TK L+ + RS +P E+F+KA+YT DIGQND+ G +
Sbjct: 373 SPFSLDIQIVQFNQFKARTKQLYQEANNSLERSKLPVPEEFAKALYTFDIGQNDLSVGFR 552
Query: 174 K----PNS 177
K PNS
Sbjct: 553 KDEF*PNS 576
Score = 30.8 bits (68), Expect = 0.79
Identities = 14/34 (41%), Positives = 23/34 (67%)
Frame = +2
Query: 171 GLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEE 204
GL K N +++R ++PDI++Q AVQK+Y +
Sbjct: 545 GLGKMNF--DQIRETMPDIVNQLASAVQKIYTNK 640
>TC83609 similar to GP|5295941|dbj|BAA81842.1 ESTs AU075322(C11109)
D22430(C11109) correspond to a region of the predicted
gene.~Similar to, partial (22%)
Length = 437
Score = 100 bits (248), Expect = 1e-21
Identities = 48/102 (47%), Positives = 68/102 (66%)
Frame = +2
Query: 1 MNTMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGS 60
MN + I++L L V +C + AI+NFGDSNSDTG YA A P G+++F
Sbjct: 125 MNILLFIFMLVLPCL-VGLSQGECDFKAIFNFGDSNSDTGGFYAAFPAESGPYGMTYFNK 301
Query: 61 ISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFA 102
+GR DGRL++DFI++ + +P+LS YL S+GS Y+HGAN+A
Sbjct: 302 PAGRASDGRLVIDFIAQAIGIPFLSPYLQSIGSYYKHGANYA 427
>TC79343 weakly similar to PIR|E86411|E86411 protein F1K23.18 [imported] -
Arabidopsis thaliana, partial (52%)
Length = 1420
Score = 85.5 bits (210), Expect = 3e-17
Identities = 60/187 (32%), Positives = 90/187 (48%), Gaps = 5/187 (2%)
Frame = +3
Query: 152 EDFSKAIYTI-DIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWI 210
E F+ +++ + +IG ND Y L S E ++ +P ++S T A+ +L + AR I
Sbjct: 594 EVFANSLFLMGEIGGNDFNYPLFIRRSIVE-IKTYVPHVISAITSAINELIDLGARTLMI 770
Query: 211 HNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAK 270
P+ C Y Y +K D+ GC+K NE A+ YN++L+ ++ +LRR+ P A
Sbjct: 771 PGNFPLGCNVIYLTKY-ETTDKSQYDSAGCLKWLNEFAEFYNQELQYELHRLRRIHPHAT 947
Query: 271 FTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY----CGKKSIKNGTVYGIAC 326
Y D Y L N GF L+ CCG G ++ CGK + AC
Sbjct: 948 IIYADYYNALLPLYQNPTKFGFTG-LKNCCG--MGGSYNFGSGSCGKPGV-------FAC 1097
Query: 327 DDSSTYI 333
DD S YI
Sbjct: 1098DDPSQYI 1118
Score = 65.1 bits (157), Expect = 4e-11
Identities = 43/99 (43%), Positives = 59/99 (59%), Gaps = 10/99 (10%)
Frame = +2
Query: 18 ACPSKKCVYPAIYNFGDSNSDTGAGY-ATMAAVEH----PNGISFFGSISGRCCDGRLIL 72
AC S Y +I++FGDS +DTG Y ++ +H P G ++F SGRC DGRLI+
Sbjct: 179 ACSS----YSSIFSFGDSIADTGNLYLSSQPPSDHCFFPPYGQTYFHHPSGRCSDGRLII 346
Query: 73 DFISEELELPYLSSYL---NSV--GSNYRHGANFAVASA 106
DFI+E L +P + YL N V ++ + GANFAV A
Sbjct: 347 DFIAESLGIPMVKPYLGIKNGVLEDNSAKEGANFAVIGA 463
>TC88477 similar to GP|21592417|gb|AAM64368.1 lipase/hydrolase putative
{Arabidopsis thaliana}, partial (92%)
Length = 1415
Score = 84.7 bits (208), Expect = 5e-17
Identities = 84/300 (28%), Positives = 131/300 (43%), Gaps = 15/300 (5%)
Frame = +2
Query: 27 PAIYNFGDSNSDTGA--GYATMAAVEH-PNGISFFGSISGRCCDGRLILDFISEELELP- 82
P + FGDS D G G ++A ++ P GI F G +GR +G+ +D I+E L
Sbjct: 179 PWYFIFGDSLVDNGNNNGLQSLARADYLPYGIDF-GGPTGRFSNGKTTVDAIAELLGFDD 355
Query: 83 YLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEP 142
Y+ Y ++ G N+A A+A IR G Q+ + F + + + ++
Sbjct: 356 YIPPYASASDDAILKGVNYASAAAGIRE------ETGRQLGARLSFSAQVQ---NYQSTV 508
Query: 143 PLRSGVPRTED-----FSKAIYTIDIGQNDI--GYGLQKPNSSEEEVRRS--IPDILSQF 193
+ TED SK IY+I +G ND Y + + ++ ++ D++ +
Sbjct: 509 SQVVNILGTEDQAASHLSKCIYSIGLGSNDYLNNYFMPQFYNTHDQYTPDEYADDLIQSY 688
Query: 194 TQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANG--CVKPHNELAQEY 251
T+ ++ LYN AR + G I C P NE A+G CV+ N Q +
Sbjct: 689 TEQLRTLYNN*ARKMVLFGIGQIGCSP---------NELATRSADGVTCVEEINSANQIF 841
Query: 252 NRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYC 311
N +LK V Q P +K YV+ Y + +ISN GF CCG + N C
Sbjct: 842 NNKLKGLVDQFNNQLPDSKVIYVNSYGIFQDIISNPSAYGFSVTNAGCCGVGRNNGQFTC 1021
>TC86678 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1341
Score = 83.2 bits (204), Expect = 1e-16
Identities = 79/298 (26%), Positives = 129/298 (42%), Gaps = 11/298 (3%)
Frame = +2
Query: 28 AIYNFGDSNSDTGAG-YATMAAVEHPN----GISFFGSISGRCCDGRLILDFISEELELP 82
A++ FGDS D G Y + N G ++F +GR DGRLI DFI+E + +P
Sbjct: 182 ALFIFGDSFLDAGNNNYINTTTFDQANFLPYGETYFNFPTGRFSDGRLISDFIAEYVNIP 361
Query: 83 YLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEP 142
+ +L + Y +G NFA A + + F+ +Q I FK T L
Sbjct: 362 LVPPFLQPDNNKYYNGVNFASGGAGALVETFQGSVIPFK-TQAINFKKVTTWL-----RH 523
Query: 143 PLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRS------IPDILSQFTQA 196
L S +T S A+Y IG ND P + +V + + ++ FT
Sbjct: 524 KLGSSDSKTL-LSNAVYMFSIGSND----YLSPFLTNSDVLKHYSHTEYVAMVIGNFTST 688
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
+++++ A+ F I N P+ C+P KG+ C++ + LA +N+ L
Sbjct: 689 IKEIHKRGAKKFVILNLPPLGCLPGTRII--QSQGKGS-----CLEELSSLASIHNQALY 847
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYCGKK 314
+ + +L++ KF+ D + +I++ GF CCGS + CG K
Sbjct: 848 EVLLELQKQLRGFKFSLYDFNSDLSHMINHPLKYGFKEGKSACCGSGPFRGEYSCGGK 1021
>TC76727 similar to GP|10638955|emb|CAB81548. putative proline-rich protein
APG isolog {Cicer arietinum}, partial (92%)
Length = 1919
Score = 83.2 bits (204), Expect = 1e-16
Identities = 80/309 (25%), Positives = 127/309 (40%), Gaps = 9/309 (2%)
Frame = -2
Query: 3 TMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAG--YATMAAVEHPN-GISFFG 59
T+ L+ + CF L ++ + PAI FGDS D G T+ +P G F
Sbjct: 1156 TLVLLIVSCF--LTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTN 983
Query: 60 SI-SGRCCDGRLILDFISEELELP-YLSSYLN--SVGSNYRHGANFAVASAPIRPIIAGL 115
+GR C+G+L DF +E L + +YL+ + G N GANFA A++ A L
Sbjct: 982 KQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATL 803
Query: 116 TYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDI--GYGLQ 173
+ Q FK + L S + ++Y + G +D Y
Sbjct: 802 NHAIPLSQQLEYFKEYQGKLAQVAGSKKAASII------KDSLYVLSAGSSDFVQNYYTN 641
Query: 174 KPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKG 233
+ V + +L FT ++ +Y AR + + P+ C+P + +
Sbjct: 640 PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHE--- 470
Query: 234 NLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFV 293
NGCV N AQ +N+++ L++ P K D+Y Y L+ N N GF
Sbjct: 469 ----NGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNFGFA 302
Query: 294 NPLEFCCGS 302
+ CCG+
Sbjct: 301 EAGKGCCGT 275
>TC89608 similar to PIR|T01143|T01143 probable GDSL-motif lipase/hydrolase
[imported] - Arabidopsis thaliana, partial (70%)
Length = 959
Score = 80.5 bits (197), Expect = 9e-16
Identities = 62/225 (27%), Positives = 94/225 (41%), Gaps = 11/225 (4%)
Frame = +2
Query: 89 NSVGSNYRHGANFAVASAPIRP----IIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPL 144
N+ G + +G N+A I I + Q+ F + + L Q
Sbjct: 17 NATGKSILYGVNYASGGGGILNATGRIFVNRIGMDIQIDYFTITRKQIDKLLGQS----- 181
Query: 145 RSGVPRTEDF--SKAIYTIDIGQNDIGYGLQKPNSS-----EEEVRRSIPDILSQFTQAV 197
+ DF K+I++I +G ND P S + + D+++ F +
Sbjct: 182 -----KARDFIMKKSIFSITVGANDFLNNYLLPVLSVGARISQSPDAFVDDMINHFRGQL 346
Query: 198 QKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKD 257
+LY +AR F I N GPI CIPY + L+ + CV N+LA +YN +LKD
Sbjct: 347 TRLYKMDARKFVIGNVGPIGCIPY-------QKTINQLNEDECVDLANKLAIQYNGRLKD 505
Query: 258 QVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGS 302
+ +L P A F +VY + LI N GF CCG+
Sbjct: 506 MLAELNDNLPGATFVLANVYDLVMELIKNYDKYGFTTSSRACCGN 640
>TC80438 similar to PIR|E96579|E96579 hypothetical protein T18A20.15
[imported] - Arabidopsis thaliana, partial (28%)
Length = 857
Score = 75.1 bits (183), Expect = 4e-14
Identities = 71/251 (28%), Positives = 112/251 (44%), Gaps = 13/251 (5%)
Frame = +2
Query: 28 AIYNFGDSNSDTGAGYATMAAVEHP--------NGISFFGSISGRCCDGRLILDFISEEL 79
A++ FGDS D+G E+ NG+ F +GR DGR+I DFI+E
Sbjct: 143 ALFIFGDSTVDSGNNNYIDTIPENKADCKPYGQNGV--FDKPTGRFSDGRVITDFIAEYA 316
Query: 80 ELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQV---SQFILFKSHTKILF 136
+LP + YL +Y +G NFA A + P T G + +Q F+ K L
Sbjct: 317 KLPLIPPYLKP-SIDYSNGVNFASGGAGVLP----ETNQGLVIDLPTQLSNFEEVRKSLA 481
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQND-IGYGLQKPNSSEE-EVRRSIPDILSQFT 194
++ E + E S+A+Y I IG ND +G L P E ++ I ++ T
Sbjct: 482 EKLGEEKAK------ELISEAVYFISIGSNDYMGGYLGNPKMQESYNPQQYIGMVIGNLT 643
Query: 195 QAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQ 254
Q++ +LY + AR F + P+ C+P P ++ GC + LA +N
Sbjct: 644 QSIVRLYEKGARKFGFLSLSPLGCLPALRAANPEASK------GGCFGAASSLALAHNNA 805
Query: 255 LKDQVFQLRRM 265
L + + L ++
Sbjct: 806 LSNILTSLNQV 838
>TC80740 similar to GP|10177228|dbj|BAB10602. GDSL-motif
lipase/hydrolase-like protein {Arabidopsis thaliana},
partial (94%)
Length = 1167
Score = 72.8 bits (177), Expect = 2e-13
Identities = 88/334 (26%), Positives = 129/334 (38%), Gaps = 13/334 (3%)
Frame = +3
Query: 2 NTMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAG---YATMAAVEHPNGISFF 58
N +T + + FN+ P + PA++ FGDS D Y + + P G F
Sbjct: 123 NFLTFLLLFVVFNVVKGQP----LVPALFIFGDSVVDARNNNNLYTIVKSNFPPYGRDFN 290
Query: 59 GSI-SGRCCDGRLILDFISEELELP-YLSSYLN--SVGSNYRHGANFAVASAPIRPIIAG 114
+ +GR C+G+L DF +E L Y +YLN N +GANFA ++ A
Sbjct: 291 NQMPTGRFCNGKLAADFTAENLGFTTYPPAYLNLQEKRKNLLNGANFASGASGYFDPTAK 470
Query: 115 LTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQ- 173
L + Q +K IL + S + S AIY + G +D
Sbjct: 471 LYHAISLEQQLEHYKECQNILVGVAGKSNASSII------SGAIYLVRAGSSDFVQNYYI 632
Query: 174 KPNSSEEEVRRSIPDILSQ-FTQAVQKLYNEEARVFWIHNTGPIECIPYYY-FFYPHKNE 231
P + DIL Q +T +Q LY AR + P+ C+P F H NE
Sbjct: 633 NPLLYKVFTADQFSDILMQHYTIFIQNLYALGARKIGVTTLPPLGCLPAAITLFGSHSNE 812
Query: 232 KGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQG 291
CV N A +N +L L++ +D+Y + L++ G
Sbjct: 813 --------CVDRLNNDALNFNTKLNTTSQNLQKELSNLTLAVLDIYQPLHDLVTKPTENG 968
Query: 292 FVNPLEFCCGSYQGNEIHYCGKKSI---KNGTVY 322
F + CCG+ C K SI N T Y
Sbjct: 969 FYEARKACCGTGLIETSILCNKDSIGTCANATEY 1070
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,869,769
Number of Sequences: 36976
Number of extensions: 174649
Number of successful extensions: 1047
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 990
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 998
length of query: 341
length of database: 9,014,727
effective HSP length: 97
effective length of query: 244
effective length of database: 5,428,055
effective search space: 1324445420
effective search space used: 1324445420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136841.9


