

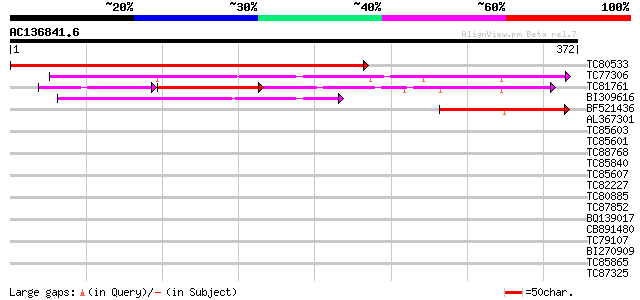
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.6 + phase: 0
(372 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80533 weakly similar to GP|6041833|gb|AAF02142.1| unknown prot... 459 e-130
TC77306 similar to GP|17064820|gb|AAL32564.1 Unknown protein {Ar... 173 1e-43
TC81761 similar to PIR|A86446|A86446 unknown protein [imported] ... 103 8e-37
BI309616 similar to GP|18086465|gb At1g48450/T1N15_5 {Arabidopsi... 106 1e-23
BF521436 similar to GP|17064820|gb| Unknown protein {Arabidopsis... 77 1e-14
AL367301 similar to GP|18086465|gb| At1g48450/T1N15_5 {Arabidops... 32 0.30
TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 32 0.30
TC85601 nodulin 30 1.1
TC88768 weakly similar to PIR|T14319|T14319 protein AX110P - car... 29 2.6
TC85840 similar to SP|P35016|ENPL_CATRO Endoplasmin homolog prec... 29 2.6
TC85607 homologue to GP|437310|gb|AAA62850.1|| nodulin {Medicago... 29 2.6
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 29 3.3
TC80885 apyrase-like protein [Medicago truncatula] 29 3.3
TC87852 similar to GP|5263329|gb|AAD41431.1| Contains PF|00646 F... 28 5.7
BQ139017 28 7.4
CB891480 weakly similar to GP|12056928|gb putative resistance pr... 28 7.4
TC79107 similar to GP|8569098|gb|AAF76443.1| ESTs gb|F20048 gb|... 28 7.4
BI270909 similar to GP|8569098|gb|A ESTs gb|F20048 gb|F20049 co... 28 7.4
TC85865 glutamine synthetase [Medicago truncatula] 28 7.4
TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Ar... 24 7.6
>TC80533 weakly similar to GP|6041833|gb|AAF02142.1| unknown protein
{Arabidopsis thaliana}, partial (41%)
Length = 735
Score = 459 bits (1182), Expect = e-130
Identities = 234/235 (99%), Positives = 234/235 (99%)
Frame = +1
Query: 1 MENCLTIKPHIGFFTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGK 60
MENCLTIKPHIGFFTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGK
Sbjct: 31 MENCLTIKPHIGFFTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGK 210
Query: 61 FLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEV 120
FLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEV
Sbjct: 211 FLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEV 390
Query: 121 AVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLDMIREHV 180
AVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLDMIREHV
Sbjct: 391 AVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLDMIREHV 570
Query: 181 TTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNL 235
TTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSL YHLERNLNL
Sbjct: 571 TTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLTYHLERNLNL 735
>TC77306 similar to GP|17064820|gb|AAL32564.1 Unknown protein {Arabidopsis
thaliana}, partial (69%)
Length = 1709
Score = 173 bits (438), Expect = 1e-43
Identities = 118/361 (32%), Positives = 193/361 (52%), Gaps = 19/361 (5%)
Frame = +3
Query: 27 RSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLAD 86
RS V S+ +S +SS PL ++ +G+FLS +L NH +L AV ++L L
Sbjct: 345 RSFVVRAATSSSSSPDPDSSHKIAPLQFQSPIGQFLSQILINHPHLVPAAVDQQLLQLQS 524
Query: 87 DRDAANSRM---LLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVP 143
DRD A+ + + S + +L+RRIAE+K N+ A+E+I+ L+ KF + L+P
Sbjct: 525 DRDVAHHQQNQDPSPTTSTDLVLYRRIAEVKANERRTALEEILYTLVVQKFMDANISLIP 704
Query: 144 KLSRCLYNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQ 203
++ +GR++ ++D +LE +H+ E +MI+ H+ + G +A + + ++ +
Sbjct: 705 SITPDA-SGRVDSWSNEDGKLEQLHSNEAYEMIQNHLALILGNRA----GDLSSVAQISK 869
Query: 204 FLLGRIYVASILYGYFLKSVSLRYHLERNLNL--ANHDVHPGHRTNLSFKDMCPYGFEDD 261
+G++Y AS++YGYFLK V R+ LE+++ + + D H+T D G E D
Sbjct: 870 LRVGQVYAASVMYGYFLKRVDQRFQLEKSMKVLTSASDDSSIHQT---IVDDARPGSEVD 1040
Query: 262 IFGHLSNMK-------PIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVR 314
S+ + + G + L+ YVM F +LQR A +RSKEAV+++
Sbjct: 1041TSQVTSHPEVSTWPGGDVSPGGFGYGIKPTRLRNYVMSFDGDTLQRYATIRSKEAVSIIE 1220
Query: 315 SYSSALF-------NSEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVY 367
++ ALF EG D+ I SF L+RLVLEAV FG FLW+ E Y+D+ Y
Sbjct: 1221KHTEALFGRPGMVITHEGGIDYSEDETIKISFGGLRRLVLEAVTFGCFLWDVESYVDSRY 1400
Query: 368 K 368
+
Sbjct: 1401R 1403
>TC81761 similar to PIR|A86446|A86446 unknown protein [imported] -
Arabidopsis thaliana, partial (68%)
Length = 1264
Score = 103 bits (258), Expect(3) = 8e-37
Identities = 68/211 (32%), Positives = 113/211 (53%), Gaps = 17/211 (8%)
Frame = +1
Query: 165 ESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVS 224
+ +H+ E +MI+ H++ V G +A + +K++ LG++Y ASI+YGYFLK V
Sbjct: 658 KXVHSSEAFEMIQSHLSLVLGERAVGPLQTIIQISKIK---LGKLYAASIMYGYFLKRVD 828
Query: 225 LRYHLERNLNLANHDVHPGHRTNLSFKDMCPYG--FEDDIFGHLSNMKPIGQGLIRQEE- 281
R+ LER++ D+ + N+SF + P ++ D L + P +G ++
Sbjct: 829 ERFQLERSVGTLPQDL---GKENISFDEPSPPNKLWDSD---SLIRIYPDDEGYYEMDDM 990
Query: 282 -------EIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALF-------NSEGFD 327
+ L+ YV + +LQR A +RSKEA++L+ + ALF + +G
Sbjct: 991 NTGDGEGKSSGLRAYVTQLDTEALQRLATVRSKEAISLIEKQTQALFGRPDIRLSGDGSI 1170
Query: 328 SVDSDDVILTSFSSLKRLVLEAVAFGSFLWE 358
+D+V+ +FS L LVLE+ AFGSFLW+
Sbjct: 1171ETTNDEVLSLTFSXLTMLVLESXAFGSFLWD 1263
Score = 46.2 bits (108), Expect(3) = 8e-37
Identities = 25/70 (35%), Positives = 46/70 (65%), Gaps = 1/70 (1%)
Frame = +3
Query: 98 ASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCL-YNGRLEI 156
+S S E L++RIAE+KE + +E+IM LI +KF E + ++PK+S N +++
Sbjct: 453 SSTSYEDSLYKRIAEIKEKEKRTTLEEIMYCLIVNKFKENKISMIPKISATSDPNEQVDS 632
Query: 157 LPSKDWELES 166
P+++++LE+
Sbjct: 633 WPNQEFKLEA 662
Score = 42.0 bits (97), Expect(3) = 8e-37
Identities = 24/77 (31%), Positives = 42/77 (54%)
Frame = +2
Query: 20 FSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQE 79
++ SFR R +++AS +S + S PL + VG+ L +LQ H +LF + +
Sbjct: 236 YNEPYSFRGRGLVVRASTDSSDNFVPSP---PLQFESPVGQLLEQILQTHPHLFLATIDQ 406
Query: 80 ELKLLADDRDAANSRML 96
+L+ L +RDA R++
Sbjct: 407 QLEKLQTERDANKERIV 457
>BI309616 similar to GP|18086465|gb At1g48450/T1N15_5 {Arabidopsis thaliana},
partial (37%)
Length = 823
Score = 106 bits (265), Expect = 1e-23
Identities = 56/188 (29%), Positives = 110/188 (57%)
Frame = +2
Query: 32 LIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAA 91
+++A + ++ SSS PL + VG+FLS +L +H +L AV+ +L+ DRD
Sbjct: 275 VVRAESSSAESSRSSSNIAPLKLESPVGQFLSQILVSHPHLMSAAVERQLEQFQTDRDGY 454
Query: 92 NSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYN 151
+ ++ + +L+RRIAE+K + A+E+I+ L+ KF + LVP ++ +
Sbjct: 455 EQKEKPSASGTDLVLYRRIAEVKAKERRKAIEEIVYTLVVQKFMDANVSLVPSIT-ANPS 631
Query: 152 GRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYV 211
G+++ PS+D +LE +H+ E ++I+ H+ + G ++ +S + ++ + +G++Y
Sbjct: 632 GQVDSWPSEDGKLEDLHSPEAYELIQSHLALLLGNRS----GDSKSVAQISKLRVGQVYA 799
Query: 212 ASILYGYF 219
AS++YGYF
Sbjct: 800 ASVMYGYF 823
>BF521436 similar to GP|17064820|gb| Unknown protein {Arabidopsis thaliana},
partial (22%)
Length = 546
Score = 76.6 bits (187), Expect = 1e-14
Identities = 40/92 (43%), Positives = 57/92 (61%), Gaps = 7/92 (7%)
Frame = +1
Query: 283 IEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNS-------EGFDSVDSDDVI 335
+ L+ Y+M F +LQR A +RSKEAV+++ +++ ALF EG + D++I
Sbjct: 4 VSRLRSYMMSFDIETLQRYATIRSKEAVSIIENHTEALFGRPEIVITPEGKINSSKDEII 183
Query: 336 LTSFSSLKRLVLEAVAFGSFLWETEDYIDNVY 367
LKRLVLEAV FGSFLW+ E Y+D+ Y
Sbjct: 184 KIRIGGLKRLVLEAVTFGSFLWDVESYVDSRY 279
>AL367301 similar to GP|18086465|gb| At1g48450/T1N15_5 {Arabidopsis
thaliana}, partial (11%)
Length = 469
Score = 32.3 bits (72), Expect = 0.30
Identities = 14/29 (48%), Positives = 21/29 (72%)
Frame = +2
Query: 297 SLQRCAKLRSKEAVNLVRSYSSALFNSEG 325
+LQR A +RSKEAV+++ ++ ALF G
Sbjct: 17 TLQRYATIRSKEAVSIIEKHTEALFGRPG 103
>TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (70%)
Length = 1183
Score = 32.3 bits (72), Expect = 0.30
Identities = 17/44 (38%), Positives = 21/44 (47%)
Frame = +1
Query: 205 LLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNL 248
L +Y + L Y L+S HL RNL NH + H TNL
Sbjct: 991 LRNHLYTSHRLRSYLLRSYLFTSHL*RNLRCTNHMLRSPHFTNL 1122
Score = 30.4 bits (67), Expect = 1.1
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = +1
Query: 205 LLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNL 248
L +Y++ L + L+S HL RNL NH + H TNL
Sbjct: 826 LRNHLYISHQLRSHQLRSYLFTSHL*RNLRCTNHMLRSPHFTNL 957
>TC85601 nodulin
Length = 1933
Score = 30.4 bits (67), Expect = 1.1
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = +1
Query: 205 LLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNL 248
L +Y++ L + L+S HL RNL NH + H TNL
Sbjct: 859 LRNHLYISHQLRSHQLRSYLFTSHL*RNLRCTNHMLRSPHFTNL 990
>TC88768 weakly similar to PIR|T14319|T14319 protein AX110P - carrot,
partial (27%)
Length = 767
Score = 29.3 bits (64), Expect = 2.6
Identities = 13/28 (46%), Positives = 18/28 (63%)
Frame = +2
Query: 153 RLEILPSKDWELESIHTLEVLDMIREHV 180
RLE+ P K WE+ S T VLD ++E +
Sbjct: 560 RLEMKPEKTWEVVSRKTQIVLDAVKESI 643
>TC85840 similar to SP|P35016|ENPL_CATRO Endoplasmin homolog precursor (GRP94
homolog). [Rosy periwinkle Madagascar periwinkle],
partial (92%)
Length = 2791
Score = 29.3 bits (64), Expect = 2.6
Identities = 19/56 (33%), Positives = 30/56 (52%), Gaps = 3/56 (5%)
Frame = -3
Query: 11 IGFFTRPISFSSSLSFRSRVPLIKASAGASSHCESS---SLNTPLLPRTQVGKFLS 63
+GFF+ S S+S S S + L S+ + SS S +T LLP+ +GK ++
Sbjct: 1037 LGFFSSSSSLSASSSSLSSLGLSVVSSSSEDSSSSSAGTSTSTSLLPQI*IGKLIN 870
>TC85607 homologue to GP|437310|gb|AAA62850.1|| nodulin {Medicago
truncatula}, partial (42%)
Length = 712
Score = 29.3 bits (64), Expect = 2.6
Identities = 16/44 (36%), Positives = 21/44 (47%)
Frame = +2
Query: 205 LLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNL 248
L +Y++ L + L S HL RNL NH + H TNL
Sbjct: 410 LRNHLYISHQLRSHQLTSYLFTSHL*RNLQCTNHMLRSPHFTNL 541
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 28.9 bits (63), Expect = 3.3
Identities = 12/47 (25%), Positives = 26/47 (54%)
Frame = -2
Query: 3 NCLTIKPHIGFFTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLN 49
+CL+I H+ + +R S SL + +PL++ A+S + + ++
Sbjct: 368 SCLSIYEHLSYRSRIFSIQGSLEYSKSLPLVEKIMRATSASQRTEIS 228
>TC80885 apyrase-like protein [Medicago truncatula]
Length = 1451
Score = 28.9 bits (63), Expect = 3.3
Identities = 33/142 (23%), Positives = 61/142 (42%), Gaps = 5/142 (3%)
Frame = +2
Query: 227 YHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQEEEIEDL 286
YH ++NL+L +H G+ ++ F D G + N + + L+ EE ED+
Sbjct: 185 YHFDQNLDL----LHIGN--DIEFVDKIKPGLS----AYADNPEQAAKSLLPLLEEAEDV 334
Query: 287 KCYVMRFHPGSLQRCA-----KLRSKEAVNLVRSYSSALFNSEGFDSVDSDDVILTSFSS 341
M HP + R +L + +A + + +F++ +V SD V
Sbjct: 335 IPEDM--HPKTPLRLGATAGLRLLNGDAAEKILQATRNMFSNRSTLNVQSDAVS------ 490
Query: 342 LKRLVLEAVAFGSFLWETEDYI 363
+++ GS++W T +YI
Sbjct: 491 ----IIDGTQEGSYMWVTVNYI 544
>TC87852 similar to GP|5263329|gb|AAD41431.1| Contains PF|00646 F-box domain.
ESTs gb|Z37267 gb|R90412 gb|Z37268 and gb|T88189 come
from this, partial (97%)
Length = 1470
Score = 28.1 bits (61), Expect = 5.7
Identities = 18/56 (32%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Frame = -1
Query: 12 GFFTRPISFSSS-LSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVL 66
G F R +S SS+ + +PL+ +AG S C SSS + PR L+ ++
Sbjct: 1122 GKFKRDVSTSSNGTKTKGVMPLLCETAGTSGLCVSSSCHPSTTPRMSSSDVLASLV 955
>BQ139017
Length = 676
Score = 27.7 bits (60), Expect = 7.4
Identities = 20/65 (30%), Positives = 29/65 (43%), Gaps = 16/65 (24%)
Frame = +3
Query: 127 SLLIFHKFSEIRAPLVPK------------LSRCLYNGR----LEILPSKDWELESIHTL 170
SL +FH F+ L P +S CL++ R + +L S IHTL
Sbjct: 24 SLFVFHTFTRGEQQLTPSTVLIPLWCGHTLVSTCLHSKRYKLAISLLRSLQGFTRMIHTL 203
Query: 171 EVLDM 175
+VLD+
Sbjct: 204 KVLDL 218
>CB891480 weakly similar to GP|12056928|gb putative resistance protein
{Glycine max}, partial (7%)
Length = 622
Score = 27.7 bits (60), Expect = 7.4
Identities = 14/41 (34%), Positives = 23/41 (55%), Gaps = 3/41 (7%)
Frame = +2
Query: 291 MRFHPGSLQRCAKLRSKEAVNLVRSYSSALF---NSEGFDS 328
+R H GS Q + K+ ++++ + +ALF N GFDS
Sbjct: 194 VRKHTGSYQTALAKQKKQGKDMIQRWKNALFEVANLSGFDS 316
>TC79107 similar to GP|8569098|gb|AAF76443.1| ESTs gb|F20048 gb|F20049 come
from this gene. {Arabidopsis thaliana}, partial (59%)
Length = 949
Score = 27.7 bits (60), Expect = 7.4
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = +3
Query: 15 TRPISFSSSLSFRS--RVPLIKASAGASSHCESSSLNTP 51
T+PISFS SLS + R L + S S+ +SSLN P
Sbjct: 135 TKPISFSLSLSLMAFLRPTLSQLSLSQSNFPRNSSLNHP 251
>BI270909 similar to GP|8569098|gb|A ESTs gb|F20048 gb|F20049 come from this
gene. {Arabidopsis thaliana}, partial (22%)
Length = 659
Score = 27.7 bits (60), Expect = 7.4
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = +3
Query: 15 TRPISFSSSLSFRS--RVPLIKASAGASSHCESSSLNTP 51
T+PISFS SLS + R L + S S+ +SSLN P
Sbjct: 102 TKPISFSLSLSLMAFLRPTLSQLSLSQSNFPRNSSLNHP 218
>TC85865 glutamine synthetase [Medicago truncatula]
Length = 1760
Score = 27.7 bits (60), Expect = 7.4
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Frame = -3
Query: 1 MENCLTIKPHIGFFTRPISFSSSL-----SFRSRVPLIKASAGASSHCESSSLNTPLLPR 55
+ENCL I H FTR +S + ++ R + + + ++H +S S N +L
Sbjct: 579 LENCLRI*DHFTIFTRGLSSARTIVVPLRKLRGMLNRL*YGS*FAAHIDSCSTNPDVLGN 400
Query: 56 TQVGKF 61
VG++
Sbjct: 399 YAVGEW 382
>TC87325 similar to GP|18176187|gb|AAL60000.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1568
Score = 23.9 bits (50), Expect(2) = 7.6
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +2
Query: 21 SSSLSFRSRVPLIKASAGASSHCESSSLNTPLL 53
S+S S R +P + A S+CE SS N LL
Sbjct: 1004 STSYSGRHNIPGLAADKFCYSYCE*SSPNVILL 1102
Score = 21.9 bits (45), Expect(2) = 7.6
Identities = 10/27 (37%), Positives = 15/27 (55%)
Frame = +1
Query: 66 LQNHRNLFHVAVQEELKLLADDRDAAN 92
L+ HR+ FHV + +K L + A N
Sbjct: 1162 LERHRHHFHVYRSQHIKYLYPRKIAEN 1242
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,747,146
Number of Sequences: 36976
Number of extensions: 165854
Number of successful extensions: 839
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 802
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 832
length of query: 372
length of database: 9,014,727
effective HSP length: 98
effective length of query: 274
effective length of database: 5,391,079
effective search space: 1477155646
effective search space used: 1477155646
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136841.6


