

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136840.7 - phase: 0 /pseudo
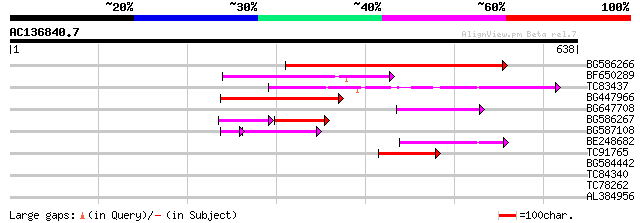
(638 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 194 1e-49
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 126 3e-29
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 113 3e-25
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 110 1e-24
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 88 1e-17
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 50 3e-14
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 69 5e-14
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 74 2e-13
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 57 2e-08
BG584442 35 0.12
TC84340 homologue to PIR|F96704|F96704 hypothetical protein T23K... 29 6.3
TC78262 similar to GP|20260620|gb|AAM13208.1 unknown protein {Ar... 29 6.3
AL384956 homologue to GP|9886727|emb PPF-1 protein {Pisum sativu... 29 6.3
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 194 bits (492), Expect = 1e-49
Identities = 103/251 (41%), Positives = 151/251 (60%), Gaps = 1/251 (0%)
Frame = -3
Query: 311 DFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRI 370
++R I+ CN YKII K+L R++P+L +II P QS+F+PGR+ +DN ++ +++HY+R
Sbjct: 772 EYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLRQ 593
Query: 371 SKKKKGY-VAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGN 429
S KK +A K D+ KA+D + W+FLR L GF I IM CV++ S+S L NG
Sbjct: 592 SGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLINGG 413
Query: 430 RLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFA 489
P+ GLRQGDPLSPYLFILC E LS A+++G+ + V+ P ++HLLFA
Sbjct: 412 PQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLFA 233
Query: 490 DDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSISGIR 549
DD + F K+N++ + + D++ SG IN KS +SS +Q I+ + I
Sbjct: 232 DDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKIA 53
Query: 550 CTTSLDKYLGF 560
KYLG+
Sbjct: 52 KEGGTGKYLGY 20
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 126 bits (316), Expect = 3e-29
Identities = 70/197 (35%), Positives = 112/197 (56%), Gaps = 3/197 (1%)
Frame = +3
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
+V AL S KAPG DG++ FFK +I+GD V + F T I+ T + L
Sbjct: 36 EVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKIINCTYVTL 215
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
+PK + K+FRPI+ C++IYKII+K+L R++ +LN+++ QS+F+ GR DN I
Sbjct: 216 LPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKGRVIFDNII 395
Query: 360 VLQEVIHYMRISKKKKGY---VAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHC 416
+ E++ S +KG KIDL KA+D+ W F++ + + GFP + +M
Sbjct: 396 LSHELVK----SYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNWVMAX 563
Query: 417 VTSSSFSILWNGNRLPP 433
+T++S++ NG+ P
Sbjct: 564 LTTASYTFNXNGDLTXP 614
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 113 bits (282), Expect = 3e-25
Identities = 95/331 (28%), Positives = 153/331 (45%), Gaps = 3/331 (0%)
Frame = +2
Query: 292 ISDTLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPG 351
I+ T IALIPKVD P DFRPISL +YKI+ K+L +RLR ++ ++I QS+F+
Sbjct: 50 INSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKN 229
Query: 352 RSTTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFD---NVNWDFLRSCLCDFGFPDT 408
R + + L ++ +MR+ +K + + L++ + W F
Sbjct: 230 RQILE-MVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWILF---*VGMSFLVL 397
Query: 409 TIKLIMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQ 468
K I CV++++ S+L NG+ PT+ +L + + +
Sbjct: 398 WRKWIKECVSTATTSVLVNGS------PTN-------------VLMKSLVQTQLFTRYSF 520
Query: 469 GSWEPIHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRA 528
G P+ V SHL FA+D LL N +R +R F SGLK+N HKS
Sbjct: 521 GVVNPVVV-------SHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKS-G 676
Query: 529 FYSSGVTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQN 588
+ ++ S+ + YLG PI + S + I+++++ RL W +
Sbjct: 677 LVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIKARLTGWNS 856
Query: 589 KLLNKPGRLALASFVLSSISTYYMQINWLPQ 619
+ L+ GRL L VL+S+S Y + + L Q
Sbjct: 857 RFLSFGGRLVLLKSVLTSLSVYALPSSKLHQ 949
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 110 bits (276), Expect = 1e-24
Identities = 58/138 (42%), Positives = 84/138 (60%)
Frame = +2
Query: 238 KKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLI 297
+++V A++ P KAPGPDG +FF++Y HIVG +V + S ++ T I
Sbjct: 263 EEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIVGKEVQQMVLQVLNNSMETEELNKTFI 442
Query: 298 ALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDN 357
LIPK P T KD+RPISLCN++ KIITKV+ +R++ L ++I QS+F+ GR TDN
Sbjct: 443 VLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRLITDN 622
Query: 358 AIVLQEVIHYMRISKKKK 375
A++ V R +K K
Sbjct: 623 ALIAWSVSIG*RXRRKGK 676
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 87.8 bits (216), Expect = 1e-17
Identities = 45/99 (45%), Positives = 56/99 (56%)
Frame = +1
Query: 436 PTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFADDVLLF 495
P GLRQGDPLSPYLFILC LS + + + I V+ PK++HLLFADD LLF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 496 TKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
+AN T+ I + + SG +N KS YS V
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNV 300
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 50.1 bits (118), Expect(2) = 3e-14
Identities = 26/62 (41%), Positives = 39/62 (61%)
Frame = +1
Query: 299 LIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNA 358
L+PK +FRPISLCN+ YKI++KVL RL+ +L II Q++F + +DN
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 359 IV 360
++
Sbjct: 781 LI 786
Score = 46.6 bits (109), Expect(2) = 3e-14
Identities = 20/62 (32%), Positives = 36/62 (57%)
Frame = +3
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
+++++V A+ P+K PGPDG + FF+Q+ +GDD+ ++A T + I+ T
Sbjct: 411 ISREEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKT 590
Query: 296 LI 297
I
Sbjct: 591 NI 596
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 68.6 bits (166), Expect(2) = 5e-14
Identities = 36/89 (40%), Positives = 51/89 (56%)
Frame = +3
Query: 263 FFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKDFRPISLCNIIY 322
FF+ HI+ D+ + S + D ++ T I LIPK P + RPISLCN+ Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 323 KIITKVLVHRLRPILNNIIGPYQSSFLPG 351
KII+KVL RL+ L ++I QS+F+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 27.3 bits (59), Expect(2) = 5e-14
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +2
Query: 238 KKKVTAALHSTKPYKAPGPDGFHCIFF 264
+++V AL P KAPGPDG + F
Sbjct: 335 EEEVRLALFIMHPEKAPGPDGMTTLLF 415
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 73.6 bits (179), Expect = 2e-13
Identities = 43/123 (34%), Positives = 62/123 (49%)
Frame = +3
Query: 439 GLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFADDVLLFTKA 498
GL+QGDPL+P+LF+L E +S + NAV + ++ V G ++SHL +ADD L
Sbjct: 27 GLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGMP 206
Query: 499 NSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSISGIRCTTSLDKYL 558
L ++ L F SGLK+N HKS + V +D + R + YL
Sbjct: 207 TVDNLWTLKALLQGFEMASGLKVNFHKS-SLIGINVPRDFMEAACRFLNCREESIPFIYL 383
Query: 559 GFP 561
G P
Sbjct: 384 GLP 392
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 57.0 bits (136), Expect = 2e-08
Identities = 27/69 (39%), Positives = 42/69 (60%)
Frame = +2
Query: 416 CVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIH 475
CV S+ + +L N + + P P+ GL+QGD LSPY+FI+C+E LS I +A ++G
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 476 VSNPGPKLS 484
+ P +S
Sbjct: 284 I*RGAPPVS 310
Score = 35.4 bits (80), Expect = 0.068
Identities = 23/87 (26%), Positives = 40/87 (45%)
Frame = +3
Query: 486 LLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSI 545
LLF L + + ++++ + + SG I+L KS + S V ++T I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 546 SGIRCTTSLDKYLGFPIIKGRPKKSDF 572
G++ KYLG P + GR + + F
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTF 539
>BG584442
Length = 775
Score = 34.7 bits (78), Expect = 0.12
Identities = 19/57 (33%), Positives = 29/57 (50%)
Frame = +1
Query: 578 KMQKRLASWQNKLLNKPGRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
K K++ +NK L+K + + L SIS+Y M I L S D I+++ F W
Sbjct: 370 KFDKKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSW 540
>TC84340 homologue to PIR|F96704|F96704 hypothetical protein T23K23.2
[imported] - Arabidopsis thaliana, partial (33%)
Length = 692
Score = 28.9 bits (63), Expect = 6.3
Identities = 11/23 (47%), Positives = 17/23 (73%)
Frame = +3
Query: 310 KDFRPISLCNIIYKIITKVLVHR 332
KD + CN+I K+I+K++VHR
Sbjct: 591 KDVPKVMQCNLIIKLISKLVVHR 659
>TC78262 similar to GP|20260620|gb|AAM13208.1 unknown protein {Arabidopsis
thaliana}, partial (93%)
Length = 3014
Score = 28.9 bits (63), Expect = 6.3
Identities = 23/84 (27%), Positives = 36/84 (42%), Gaps = 7/84 (8%)
Frame = +3
Query: 555 DKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLN-------KPGRLALASFVLSSI 607
D YL ++ K F+ K+Q L +QNKL K G L L + +I
Sbjct: 972 DNYLTHQVVYNANKLEKFVKKKSKLQNWLVYYQNKLERTSKRPEMKTGFLGLHGKKVDAI 1151
Query: 608 STYYMQINWLPQSVCDNIDQVTRN 631
Y +I+ L + + D+VT +
Sbjct: 1152 DYYTTEIDKLSKEIALERDKVTND 1223
>AL384956 homologue to GP|9886727|emb PPF-1 protein {Pisum sativum}, partial
(26%)
Length = 354
Score = 28.9 bits (63), Expect = 6.3
Identities = 18/43 (41%), Positives = 25/43 (57%), Gaps = 6/43 (13%)
Frame = +1
Query: 393 WDFLRSCLCDFGFPDTTIKLIMHC------VTSSSFSILWNGN 429
W+FL +C G+P +T L+ HC +S SFSI +NGN
Sbjct: 178 WNFLAFSIC--GWP-STFGLV*HCSVSCFTCSSYSFSICFNGN 297
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.350 0.156 0.550
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,255,595
Number of Sequences: 36976
Number of extensions: 344085
Number of successful extensions: 3143
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1771
Number of HSP's successfully gapped in prelim test: 155
Number of HSP's that attempted gapping in prelim test: 1339
Number of HSP's gapped (non-prelim): 1990
length of query: 638
length of database: 9,014,727
effective HSP length: 102
effective length of query: 536
effective length of database: 5,243,175
effective search space: 2810341800
effective search space used: 2810341800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136840.7


