

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.9 + phase: 0 /pseudo
(261 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
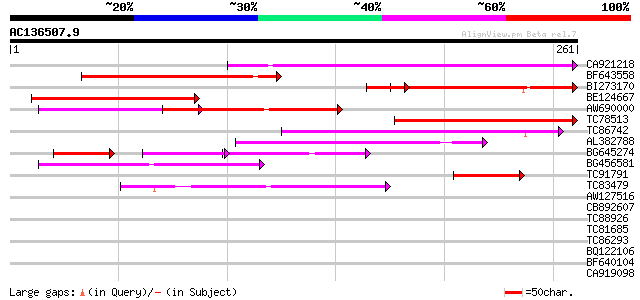
Score E
Sequences producing significant alignments: (bits) Value
CA921218 139 8e-34
BF643558 102 2e-22
BI273170 92 9e-22
BE124667 94 5e-20
AW690000 85 2e-17
TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical prot... 70 1e-12
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 67 5e-12
AL382788 weakly similar to GP|11322386|emb glycine receptor beta... 63 1e-10
BG645274 36 1e-08
BG456581 48 4e-06
TC91791 weakly similar to GP|7769858|gb|AAF69536.1| F12M16.20 {A... 47 1e-05
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 43 1e-04
AW127516 31 0.42
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 31 0.42
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 30 0.94
TC81685 29 2.1
TC86293 homologue to GP|5305145|emb|CAB46084.1 fructose-1 6-bisp... 29 2.1
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 28 2.7
BF640104 28 3.6
CA919098 28 3.6
>CA921218
Length = 707
Score = 139 bits (351), Expect = 8e-34
Identities = 67/161 (41%), Positives = 96/161 (59%)
Frame = -3
Query: 101 PIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAIN 160
P P WI NT+G+ N + +CGG FR+ N F+ F +N +A +AKL G + AI
Sbjct: 594 PEPLWINCNTNGSANINTS--ACGGTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIE 421
Query: 161 IAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNS 220
IA NW ++WLE D L V AFK+ +++ W+LRN+W N + SMNF ++H++ EGN
Sbjct: 420 IAAARNWSHLWLELDSSLVVNAFKSKSVIHWNLRNRWNNCLFLISSMNFFVSHVFREGNQ 241
Query: 221 CADCMANVGLTLSSFVWLPSLPDCIKHDYGRNRLGMHSFRF 261
CAD +AN GL+L + +P I Y N++G FRF
Sbjct: 240 CADGLANFGLSLDHLTYWNHVPPFIHRFYVENKIGWPMFRF 118
>BF643558
Length = 645
Score = 102 bits (253), Expect = 2e-22
Identities = 49/92 (53%), Positives = 62/92 (67%)
Frame = +2
Query: 34 RNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFVFLKAFKINIHPPRAPLI 93
RNQ RF +K HWKS+I +II + L+GN T + N+ +FVFLK F I+IHPP+AP I
Sbjct: 365 RNQRRFNNKMIHWKSSITSIISSTFLSGNYTSACDYTNITDFVFLKKFSIDIHPPKAPKI 544
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGG 125
EV+W PP WIK NTDG + N +SCGG
Sbjct: 545 IEVLWNPPPLHWIKCNTDG--SSNTNTSSCGG 634
>BI273170
Length = 391
Score = 92.0 bits (227), Expect(2) = 9e-22
Identities = 42/87 (48%), Positives = 61/87 (69%), Gaps = 1/87 (1%)
Frame = +1
Query: 176 LQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANVGLTLSSF 235
L L + FKN++ VPW LRN+W N R+MNF+++H++ EGN CAD +AN+GL+L+
Sbjct: 46 LHLLSMPFKNISQVPWKLRNRWENCILATRNMNFIVSHVFREGNECADMLANIGLSLNCL 225
Query: 236 -VWLPSLPDCIKHDYGRNRLGMHSFRF 261
+WL LPDCIK + +N+LG + RF
Sbjct: 226 TIWL-ELPDCIKSIFIKNKLGWPNNRF 303
Score = 28.5 bits (62), Expect(2) = 9e-22
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = +3
Query: 165 NNWRNIWLETDLQLAVLAFK 184
+NW+++WLE+D L V AF+
Sbjct: 12 HNWQSLWLESDSALVVNAFQ 71
>BE124667
Length = 540
Score = 94.0 bits (232), Expect = 5e-20
Identities = 46/77 (59%), Positives = 55/77 (70%)
Frame = -1
Query: 11 WTPQCKVVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFV 70
WTPQCKVV+ A IIN+L+ IWF RN RF+DK WK+AI+ II VSL+GN TK TS
Sbjct: 231 WTPQCKVVITACIINLLNVIWFRRNNIRFQDKVIDWKTAINMIISKVSLSGNLTKKTSAA 52
Query: 71 NMDEFVFLKAFKINIHP 87
NM EF KA K++I P
Sbjct: 51 NMLEFTIFKACKVSIKP 1
>AW690000
Length = 652
Score = 85.1 bits (209), Expect = 2e-17
Identities = 38/83 (45%), Positives = 56/83 (66%)
Frame = +3
Query: 71 NMDEFVFLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDH 130
++ F+ LK F + IHPP+AP I EV+W PPIP WIK NTDG+ + ++CGG+FR+H
Sbjct: 330 SISNFLILKKFNVTIHPPKAPKIIEVIWRPPIPHWIKCNTDGSSRSH--SSACGGIFRNH 503
Query: 131 NNIFIGAFVQNLNTNSAFNAKLL 153
+ + F +N +AF+A+LL
Sbjct: 504 DTDLLLCFAENTGECNAFHAELL 572
Score = 52.0 bits (123), Expect = 2e-07
Identities = 27/76 (35%), Positives = 41/76 (53%)
Frame = +1
Query: 14 QCKVVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMD 73
Q V+ A +INI++++WF RN RF +KK HWKS++ +I +L N L +
Sbjct: 160 QIMEVITATMINIMNSVWFARN*LRFSNKKIHWKSSLSTVISNTALFDNSQML*HLLAFL 339
Query: 74 EFVFLKAFKINIHPPR 89
F FL+ + PR
Sbjct: 340 TFSFLRNSMLLFTLPR 387
>TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical protein {Galega
orientalis}, partial (20%)
Length = 765
Score = 69.7 bits (169), Expect = 1e-12
Identities = 34/84 (40%), Positives = 51/84 (60%)
Frame = +2
Query: 178 LAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANVGLTLSSFVW 237
L VL K+ N +PW+L N+W N ++ + +I+HIY EGN AD +AN GL+L S +
Sbjct: 404 LVVLPSKSTNQIPWNL*NRWNNVKVILQGLKCIISHIYREGNQVADTLANHGLSLDSIHF 583
Query: 238 LPSLPDCIKHDYGRNRLGMHSFRF 261
+P+ + Y RN+ G +FRF
Sbjct: 584 WNDVPEYTRASYFRNKDGWSNFRF 655
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 67.4 bits (163), Expect = 5e-12
Identities = 45/132 (34%), Positives = 61/132 (46%), Gaps = 2/132 (1%)
Frame = -1
Query: 126 VFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKN 185
V RD F+GA N+ + A+ + AI A E NI LETD V AF
Sbjct: 468 VIRDSQFGFLGALSCNIGHATPLEAEFCACMIAIEKAMELGLNNICLETDSLKVVNAFHK 289
Query: 186 LNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANVGLTLSSFV--WLPSLPD 243
+ +PW +R +W N S+ + HI EGN AD +A G LS F W P+ P
Sbjct: 288 IVGIPWQMRVRWHNCIRFCHSIACVCVHIPREGNLVADALARHGQGLSLFFLQWWPAPPS 109
Query: 244 CIKHDYGRNRLG 255
I+ ++R G
Sbjct: 108 FIQSFLAQDRYG 73
>AL382788 weakly similar to GP|11322386|emb glycine receptor betaZ subunit
{Danio rerio}, partial (4%)
Length = 353
Score = 62.8 bits (151), Expect = 1e-10
Identities = 40/116 (34%), Positives = 50/116 (42%)
Frame = +2
Query: 105 WIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGE 164
WIK D A + + G +FRD G F+ N A +A+ A+ I
Sbjct: 11 WIKAKIDNATCGSSDPSIVGSLFRDQFGSNFGCFIIFFGVNFAIHAEFYAAAYAVEIVYL 190
Query: 165 NNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNS 220
NW N WL DL+L V AF + V WSL + MNF HIY E NS
Sbjct: 191 CNWHNFWLACDLKLVVDAFHSNYKVLWSLAT*FF-----CNQMNF*AFHIYKERNS 343
>BG645274
Length = 641
Score = 36.2 bits (82), Expect(3) = 1e-08
Identities = 17/40 (42%), Positives = 22/40 (54%)
Frame = -1
Query: 62 NRTKLTSFVNMDEFVFLKAFKINIHPPRAPLIKEVMWIPP 101
+ T S+ NM EF L+ F + H P A IK V+W PP
Sbjct: 485 DHTNAHSYSNM*EFEILRVFFV*DHAPNASRIKAVLWYPP 366
Score = 35.8 bits (81), Expect(3) = 1e-08
Identities = 20/68 (29%), Positives = 35/68 (51%)
Frame = -3
Query: 99 IPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITA 158
+ PI W+K N DG++ A+C G+FRD + A N++ +F A+ ++
Sbjct: 375 VSPILSWVKCNIDGSIKGCFRPATCSGIFRDEDA*I*VAV--NMHNTPSFEAEFHDLMYE 202
Query: 159 INIAGENN 166
I I+ + N
Sbjct: 201 IEISYKKN 178
Score = 22.7 bits (47), Expect(3) = 1e-08
Identities = 10/28 (35%), Positives = 17/28 (60%)
Frame = -2
Query: 21 AVIINILSAIWFIRNQARFKDKKNHWKS 48
+ IIN+++ IW RN+ F K + K+
Sbjct: 607 SAIINVINIIW*CRNKQLFNGFKVNSKT 524
>BG456581
Length = 683
Score = 47.8 bits (112), Expect = 4e-06
Identities = 28/104 (26%), Positives = 47/104 (44%)
Frame = +2
Query: 14 QCKVVVKAVIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMD 73
Q + + A +++++ +IW+ RN RF K + + + L+G + T
Sbjct: 356 QVRDLYVAAVVHMVHSIWWARNNVRFSSAKVSAHAVQVRVHALIGLSGAVS--TGKCIAA 529
Query: 74 EFVFLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKN 117
+ L F+I H + V W PP PW+K NTDG+ N
Sbjct: 530 DAAILDVFRIPPHRRSMXEMVSVCWKPPSAPWVKGNTDGSXLNN 661
>TC91791 weakly similar to GP|7769858|gb|AAF69536.1| F12M16.20 {Arabidopsis
thaliana}, partial (4%)
Length = 576
Score = 46.6 bits (109), Expect = 1e-05
Identities = 20/33 (60%), Positives = 27/33 (81%)
Frame = -2
Query: 205 RSMNFLITHIYIEGNSCADCMANVGLTLSSFVW 237
R+MN ++THIY EGN AD +AN GLTL+S+V+
Sbjct: 575 RNMNCIVTHIYKEGNQVADSLANFGLTLNSYVF 477
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein
{Plasmodium falciparum 3D7}, partial (0%)
Length = 1222
Score = 42.7 bits (99), Expect = 1e-04
Identities = 35/125 (28%), Positives = 55/125 (44%), Gaps = 1/125 (0%)
Frame = +2
Query: 52 NIIVAVSLTGNRTK-LTSFVNMDEFVFLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNT 110
NI +S T R K + N+++ NI P + I W P W K+NT
Sbjct: 857 NIPYVLSFTQERIKGMVLLKNLNQIA-------NILNPVSRSIIWCEWKKPEIGWTKLNT 1015
Query: 111 DGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNI 170
DG++ K A GG+ RD+ I AFV F +L + + ++ ++I
Sbjct: 1016DGSVNKET--AGFGGLLRDYRGEPICAFVSKAPQGDTFLVELWAIWRGLVLSLGLGIKSI 1189
Query: 171 WLETD 175
W+E+D
Sbjct: 1190WVESD 1204
>AW127516
Length = 379
Score = 31.2 bits (69), Expect = 0.42
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +3
Query: 204 VRSMNFLITHIYIEGNSCADCMANVGLTLSS 234
+ S +F I H EGN CAD MA +G + +S
Sbjct: 216 IASRSFTIQHTLREGNHCADYMAKLGASSNS 308
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 31.2 bits (69), Expect = 0.42
Identities = 17/72 (23%), Positives = 31/72 (42%)
Frame = +1
Query: 99 IPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITA 158
IPP + + D + A+CGG+ +D F+ + L + S A+L G+
Sbjct: 556 IPPNSCEVALKCDYVVLNCDLNAACGGLIQDDQGHFVFHYANKLGSCSVLQAELWGI*HG 735
Query: 159 INIAGENNWRNI 170
++I + I
Sbjct: 736 LSIDWNRGYSKI 771
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 30.0 bits (66), Expect = 0.94
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = +3
Query: 108 VNTDGALTKNPT--KASCGGVFRDHNNIFIGAFVQNLNTN 145
+N DG+L + A CGGV D + ++ F Q LN N
Sbjct: 348 LNVDGSLLREREVPSAGCGGVLSDSSGKWLCGFAQKLNPN 467
>TC81685
Length = 1706
Score = 28.9 bits (63), Expect = 2.1
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -2
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGV 126
V W P W K+N+DG + + +A CGG+
Sbjct: 178 VSWTLPQSDWDKINSDG-MCQGSLRAGCGGL 89
>TC86293 homologue to GP|5305145|emb|CAB46084.1 fructose-1 6-bisphosphatase
{Pisum sativum}, complete
Length = 1377
Score = 28.9 bits (63), Expect = 2.1
Identities = 22/99 (22%), Positives = 41/99 (41%)
Frame = +1
Query: 131 NNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVP 190
NNI +G N A AKL+G+ N+ GE + L ++ L +
Sbjct: 280 NNIVLGCKFVCSAVNKAGLAKLIGLAGETNVQGEEQKKLDVLSNEVFCKALISSGRTCIL 459
Query: 191 WSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMANVG 229
S ++ + E + + + ++G+S DC ++G
Sbjct: 460 VSEEDEDAIFVEPKQRGKYCVVFDPLDGSSNIDCGVSIG 576
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 28.5 bits (62), Expect = 2.7
Identities = 34/147 (23%), Positives = 58/147 (39%), Gaps = 12/147 (8%)
Frame = -2
Query: 108 VNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL-NTNSAFNAKLLGVITAINIAGENN 166
+N DG+ NP G+ R+ +F F N+ NT+ A+L + + + +
Sbjct: 539 LNVDGSCLGNP*PTGFNGLIRNIAGLFNSGFPGNITNTSDILLAELHAIFQGLRMISDMG 360
Query: 167 WRNIWLETDLQLAVLAFKNLNMVPWSLRN----KWLNYFEHVRSMNFLIT-------HIY 215
+ V F +L+ V SL N K+ Y ++ + L+ H
Sbjct: 359 ISDF---------VCYFDSLHYV--SLINGPSMKFHVYATLIQDIKDLVITSKASVFHTL 213
Query: 216 IEGNSCADCMANVGLTLSSFVWLPSLP 242
EGN CAD + +G S + + P
Sbjct: 212 CEGNYCADFLEMLGAASDSVLTIHVSP 132
>BF640104
Length = 344
Score = 28.1 bits (61), Expect = 3.6
Identities = 23/81 (28%), Positives = 33/81 (40%), Gaps = 1/81 (1%)
Frame = -1
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL-NTNSAFNAKLLG 154
V WI P IK+N D LT G + R+ N I + + N A+ G
Sbjct: 344 VKWIKPHQGVIKINCDANLTSEDV-WGIGVITRNDNGIVMASGTWNRPGFMCPITAEAWG 168
Query: 155 VITAINIAGENNWRNIWLETD 175
V A A + ++N+ E D
Sbjct: 167 VYQAALFALDQGFQNVLFEND 105
>CA919098
Length = 776
Score = 28.1 bits (61), Expect = 3.6
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +2
Query: 105 WIKVNTDGALTKNPTKASCGGVFRDH 130
W+ +N D A++ KA+CG V DH
Sbjct: 278 WLMLNCDVAMSHLAGKAACGCVLHDH 355
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,310,331
Number of Sequences: 36976
Number of extensions: 161751
Number of successful extensions: 1160
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1135
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1152
length of query: 261
length of database: 9,014,727
effective HSP length: 94
effective length of query: 167
effective length of database: 5,538,983
effective search space: 925010161
effective search space used: 925010161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136507.9


