

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.3 + phase: 0 /pseudo
(491 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
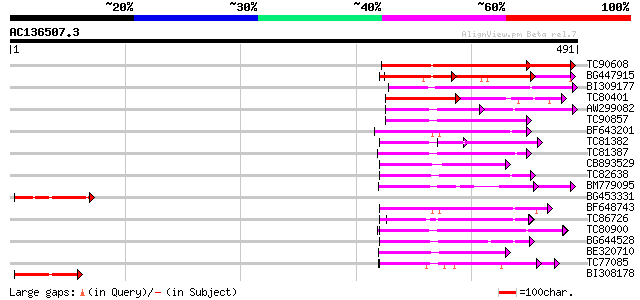
Score E
Sequences producing significant alignments: (bits) Value
TC90608 weakly similar to GP|10716599|gb|AAG21897.1 putative dis... 179 3e-45
BG447915 similar to GP|7268809|emb leucine rich repeat-like prot... 101 6e-22
BI309177 weakly similar to GP|3894383|gb|A disease resistance pr... 98 8e-21
TC80401 weakly similar to GP|17385693|dbj|BAB78644. putative Cf2... 68 9e-12
AW299082 weakly similar to GP|10177183|d receptor protein kinase... 65 6e-11
TC90857 weakly similar to GP|16930691|gb|AAL32011.1 AT4g26540/M3... 63 2e-10
BF643201 weakly similar to PIR|T10725|T107 protein kinase Xa21 (... 62 5e-10
TC81382 weakly similar to GP|14269077|gb|AAK58011.1 verticillium... 62 7e-10
TC81387 weakly similar to PIR|T45645|T45645 receptor kinase-like... 61 9e-10
CB893529 weakly similar to GP|8778255|gb| F12K21.25 {Arabidopsis... 60 1e-09
TC82638 weakly similar to PIR|T00712|T00712 protein kinase homol... 60 1e-09
BM779095 weakly similar to PIR|B84431|B8 probable receptor prote... 60 2e-09
BG453331 weakly similar to GP|20521202|dbj putative Cf2/Cf5 dise... 59 3e-09
BF648743 weakly similar to PIR|H84632|H846 probable receptor-lik... 59 4e-09
TC86726 similar to GP|14626935|gb|AAK70805.1 leucine-rich repeat... 59 4e-09
TC80900 weakly similar to PIR|G84652|G84652 probable receptor-li... 59 4e-09
BG644528 weakly similar to GP|21391894|g systemin receptor SR160... 57 1e-08
BE320710 weakly similar to GP|16924044|gb| Putative protein kina... 57 1e-08
TC77085 similar to GP|21536600|gb|AAM60932.1 putative disease re... 57 1e-08
BI308178 weakly similar to GP|6714444|gb|A putative disease resi... 57 1e-08
>TC90608 weakly similar to GP|10716599|gb|AAG21897.1 putative disease
resistance protein (3' partial) {Oryza sativa}, partial
(6%)
Length = 1494
Score = 179 bits (453), Expect = 3e-45
Identities = 96/169 (56%), Positives = 124/169 (72%), Gaps = 1/169 (0%)
Frame = +1
Query: 323 IKSNSLEGGIPKSFG-SLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQ 381
I+SNSLEGGIPKSF + C L+SL LS N+ S +L V++ +L CA+ SL+EL L+ NQ
Sbjct: 934 IRSNSLEGGIPKSFWMNACKLKSLTLSKNRFSGELQVIIDHLPK-CARYSLRELDLSFNQ 1110
Query: 382 IIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNM 441
I GT PD+S FS LE + +N L+G I ++ FP +L L + SN ++GVI++ HF M
Sbjct: 1111 INGTQPDLSIFSLLEIFDISKNRLSGKIYEDIRFPTKLRTLRMGSNSMNGVISEFHFSGM 1290
Query: 442 SMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPKWIRSQK 490
SMLK L LS NSLAL+F+ENWVPPFQL TI L SC LGP+FPKWI++QK
Sbjct: 1291 SMLKDLDLSGNSLALRFNENWVPPFQLDTIGLGSCILGPTFPKWIKTQK 1437
Score = 109 bits (273), Expect = 2e-24
Identities = 60/131 (45%), Positives = 88/131 (66%), Gaps = 1/131 (0%)
Frame = +1
Query: 323 IKSNSLEGGIPKSFGSLCSLRSLDLS-SNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQ 381
+ N L+GG+ KSF ++C+L SLDLS N L+EDL ++L NLS GC +NSL+ L ++ N+
Sbjct: 628 LSGNRLKGGVFKSFMNVCTLSSLDLSRQNNLTEDLQIILQNLSSGCVRNSLQVLDISYNE 807
Query: 382 IIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNM 441
I GT+PD+S F+SL+ + L N L+G I + S+ P++L + SN L+G I S + N
Sbjct: 808 IAGTLPDLSIFTSLKTLDLSSNQLSGKIPEGSSLPFQLEYFDIRSNSLEGGIPKSFWMNA 987
Query: 442 SMLKYLSLSSN 452
LK L+LS N
Sbjct: 988 CKLKSLTLSKN 1020
Score = 32.3 bits (72), Expect = 0.42
Identities = 21/58 (36%), Positives = 29/58 (49%)
Frame = +3
Query: 86 SFALAIS*SFRQWP*GYNPSLTWKSLSFAVP*S*FK*SCGNRSSSTWKSFKFAGTSSW 143
SFA I SF + ++ S TW+SL A+ *S + S STWK+ + SW
Sbjct: 9 SFAFEILESFVESSRWFDSSSTWRSLQLAIS*SQLQFFGRKHSISTWKACEPTRALSW 182
>BG447915 similar to GP|7268809|emb leucine rich repeat-like protein
{Arabidopsis thaliana}, partial (4%)
Length = 669
Score = 101 bits (252), Expect = 6e-22
Identities = 58/138 (42%), Positives = 88/138 (63%), Gaps = 3/138 (2%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP---VMLHNLSVGCAKNSLKELYL 377
L + N + G +P + SL++L LSSN+L+ ++P +L NLS GC +NSL+ L L
Sbjct: 159 LDLSDNRITGTLP-DLSAFTSLKTLYLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDL 335
Query: 378 ASNQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
+ N+I GT+PD+S F+SL+ + L N L+G I S+ PY+L +L + SN L+GVI S
Sbjct: 336 SDNRITGTLPDLSAFTSLKTLDLSSNQLSGEIPGGSSLPYQLEHLSITSNTLEGVIPKSF 515
Query: 438 FGNMSMLKYLSLSSNSLA 455
+ N LK L +S+NS +
Sbjct: 516 WMNACKLKSLKMSNNSFS 569
Score = 92.4 bits (228), Expect = 3e-19
Identities = 67/180 (37%), Positives = 98/180 (54%), Gaps = 14/180 (7%)
Frame = +3
Query: 325 SNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP---VMLHNLSVGCAKNSLKELYLASNQ 381
SN + G +P + SL++LDLSSN+L+ ++P +L NLS GC +NSL+ L L+ N+
Sbjct: 3 SNGIIGTLP-DLSAFTSLKTLDLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDLSDNR 179
Query: 382 IIGTVPDMSGFSSLENMFLYENLLNG------TILKN---STFPYRLANLYLDSNDLDGV 432
I GT+PD+S F+SL+ ++L N L G TIL+N L L L N + G
Sbjct: 180 ITGTLPDLSAFTSLKTLYLSSNQLTGEIPGLQTILRNLSSGCVRNSLQVLDLSDNRITGT 359
Query: 433 ITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPK--WIRSQK 490
+ D + LK L LSSN L+ + P+QL + + S TL PK W+ + K
Sbjct: 360 LPD--LSAFTSLKTLDLSSNQLSGEIPGGSSLPYQLEHLSITSNTLEGVIPKSFWMNACK 533
Score = 62.4 bits (150), Expect = 4e-10
Identities = 36/68 (52%), Positives = 49/68 (71%), Gaps = 1/68 (1%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFG-SLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
L I SN+LEG IPKSF + C L+SL +S+N S +L V++H+LS CA SL+EL L+
Sbjct: 468 LSITSNTLEGVIPKSFWMNACKLKSLKMSNNSFSGELQVIIHHLSX-CAXYSLQELDLSX 644
Query: 380 NQIIGTVP 387
N+I GT+P
Sbjct: 645 NKINGTLP 668
>BI309177 weakly similar to GP|3894383|gb|A disease resistance protein
{Lycopersicon esculentum}, partial (5%)
Length = 620
Score = 97.8 bits (242), Expect = 8e-21
Identities = 60/164 (36%), Positives = 92/164 (55%), Gaps = 1/164 (0%)
Frame = +2
Query: 329 EGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTVPD 388
+G IPKS SL L +L LS+N+L+E +P L +LK L LA N G++P
Sbjct: 2 KGQIPKSLLSLRKLETLRLSNNELNESIPDWLGQ------HENLKYLGLAENMFRGSIPS 163
Query: 389 MSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLKYL 447
G SSL ++ + + L G I + + L +L + + L GV+++ HF N+S L+ L
Sbjct: 164 SLGKLSSLVDLSVSSDFLTGNIPTSIGKLFNLKSLVIGGSSLSGVLSEIHFSNLSSLETL 343
Query: 448 SLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPKWIRSQKN 491
LS+ ++ W+PPFQL+ I L + LGP FP WI +Q++
Sbjct: 344 VLSA-PISFDMDSKWIPPFQLNGISLSNTILGPKFPTWIYTQRS 472
>TC80401 weakly similar to GP|17385693|dbj|BAB78644. putative Cf2/Cf5 disease
resistance protein homolog {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 1481
Score = 67.8 bits (164), Expect = 9e-12
Identities = 35/65 (53%), Positives = 47/65 (71%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N L+G KSF ++C+LRSL+L N +EDL +L NLS GC +NSL+ L L+SN IIGT
Sbjct: 1239 NKLKGVAFKSFMNVCTLRSLNLIQNNFTEDLQTILRNLSSGCVRNSLQVLDLSSNGIIGT 1418
Query: 386 VPDMS 390
+P +S
Sbjct: 1419 LP*LS 1433
Score = 49.3 bits (116), Expect = 3e-06
Identities = 54/168 (32%), Positives = 75/168 (44%), Gaps = 11/168 (6%)
Frame = +3
Query: 326 NSLEGG-IPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIG 384
N+ EG IP FGSL +L LDLS +P+ L +LS LK L L+ N + G
Sbjct: 393 NNFEGNSIPGFFGSLRNLTYLDLSYCYFRGQIPIQLESLS------HLKYLNLSYNLLDG 554
Query: 385 TVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG---- 439
+P G S+L+ + L N L G+I L LYL +D + H G
Sbjct: 555 LIPHRLGDLSNLQFLDLSNNYLEGSIPSQLGKLTNLQELYLSGLTID---NEDHNGGQWL 725
Query: 440 -NMSMLKYLSLSSNSLALKFSENWVPPF----QLSTIYLRSCTLGPSF 482
N++ L +L + S S L S +W+ L + LR C L F
Sbjct: 726 SNLTSLTHLHMWSIS-NLNKSNSWLKMVGKLPNLRELSLRYCDLSDHF 866
Score = 37.4 bits (85), Expect(2) = 0.001
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = +3
Query: 42 CAWEGIGCRNQTGHVEILDLNSD 64
C W+GIGC N TGHV +LDL+ +
Sbjct: 225 CQWKGIGCSNVTGHVIMLDLHGN 293
Score = 22.3 bits (46), Expect(2) = 0.001
Identities = 12/32 (37%), Positives = 17/32 (52%), Gaps = 9/32 (28%)
Frame = +1
Query: 8 CIEKERHALLELKS---------GLVLDDTYL 30
CI+ ER ALL+ K+ ++LDD L
Sbjct: 127 CIQSERQALLQFKAWPY**I*QHAVILDDRRL 222
>AW299082 weakly similar to GP|10177183|d receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (16%)
Length = 565
Score = 65.1 bits (157), Expect = 6e-11
Identities = 53/167 (31%), Positives = 78/167 (45%), Gaps = 1/167 (0%)
Frame = +2
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N L G IP G+ SLR++DLS N LS +P+ L +L L+E ++ N + G+
Sbjct: 8 NGLVGAIPNEIGNCSSLRNIDLSLNSLSGTIPLSLGSLL------ELEEFMISDNNVSGS 169
Query: 386 VP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSML 444
+P +S +L+ + + N L+G I L + N L+G I S GN S L
Sbjct: 170 IPATLSNAENLQQLQVDTNQLSGLIPPEIGKLSNLLVFFAWQNQLEGSIPSS-LGNCSKL 346
Query: 445 KYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPKWIRSQKN 491
+ L LS NSL L+ + L S + S P I S K+
Sbjct: 347 QALDLSRNSLTGSIPSRLFQLQNLTKLLLISNDISGSIPSEIGSCKS 487
Score = 51.6 bits (122), Expect = 7e-07
Identities = 33/87 (37%), Positives = 47/87 (53%), Gaps = 1/87 (1%)
Frame = +2
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N LEG IP S G+ L++LDLS N L+ +P L L +L +L L SN I G+
Sbjct: 296 NQLEGSIPSSLGNCSKLQALDLSRNSLTGSIPSRLFQL------QNLTKLLLISNDISGS 457
Query: 386 VP-DMSGFSSLENMFLYENLLNGTILK 411
+P ++ SL + L N + G+I K
Sbjct: 458 IPSEIGSCKSLIRLRLGNNRITGSIPK 538
>TC90857 weakly similar to GP|16930691|gb|AAL32011.1 AT4g26540/M3E9_30
{Arabidopsis thaliana}, partial (22%)
Length = 829
Score = 63.2 bits (152), Expect = 2e-10
Identities = 43/128 (33%), Positives = 65/128 (50%), Gaps = 1/128 (0%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
NS+ G IPK+FG+L L+ L LS N++S ++P L N L + + +N I GT
Sbjct: 147 NSITGSIPKTFGNLTLLQELQLSVNQISGEIPAELGNC------QQLTHVEIDNNLITGT 308
Query: 386 VP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSML 444
+P ++ +L +FL+ N L G I + L + L N L G I F ++
Sbjct: 309 IPSELGNLGNLTLLFLWHNKLQGNIPSTLSNCQNLEAIDLSQNLLTGPIPKGIFQLQNLN 488
Query: 445 KYLSLSSN 452
K L LS+N
Sbjct: 489 KLLLLSNN 512
Score = 30.4 bits (67), Expect = 1.6
Identities = 23/66 (34%), Positives = 32/66 (47%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + SN +EG IP+ +L LDL SN ++ LP L L SL+ L + N
Sbjct: 636 LDLGSNRIEGIIPEKISGCRNLTFLDLHSNYIAGTLPDSLSELV------SLQFLDFSDN 797
Query: 381 QIIGTV 386
I G +
Sbjct: 798 MIEGAL 815
>BF643201 weakly similar to PIR|T10725|T107 protein kinase Xa21 (EC 2.7.1.-)
A1 receptor type - long-staminate rice, partial (4%)
Length = 544
Score = 62.0 bits (149), Expect = 5e-10
Identities = 52/155 (33%), Positives = 78/155 (49%), Gaps = 19/155 (12%)
Frame = +3
Query: 317 S*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS-----VGCAKN- 370
S* LI + + G IP SFG+ +L +DLSSNKL+ +PV + N+ + +KN
Sbjct: 39 S*TRLICQEMNFVGRIPVSFGNFQNLLYMDLSSNKLNGSIPVEILNIPTLSNVLNLSKNL 218
Query: 371 ------------SLKELYLASNQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPY 417
++ + ++NQ+ G +P S SLE MFL +N+L+G I K
Sbjct: 219 LSGPIPEVGQLTTISTIDFSNNQLYGNIPSSFSNCLSLEKMFLSQNMLSGYIPKALGDVK 398
Query: 418 RLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSN 452
L L L SN L G I N+ +L+ L++S N
Sbjct: 399 GLETLDLSSNLLSGPI-PIELQNLHVLQLLNISYN 500
>TC81382 weakly similar to GP|14269077|gb|AAK58011.1 verticillium wilt
disease resistance protein Ve2 {Lycopersicon
esculentum}, partial (16%)
Length = 1974
Score = 61.6 bits (148), Expect = 7e-10
Identities = 37/92 (40%), Positives = 55/92 (59%), Gaps = 1/92 (1%)
Frame = +3
Query: 371 SLKELYLASNQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLD 430
SL LYL++N++ G++ S +S LE++ LY N L G I ++ L NL L NDL
Sbjct: 27 SLVYLYLSNNRLTGSISATSSYS-LESLNLYNNKLQGNIPESIFNLTNLTNLILSLNDLS 203
Query: 431 GVITDSHFGNMSMLKYLSLSSNS-LALKFSEN 461
G + HF ++ L++LSLS N+ L+L F N
Sbjct: 204 GFVNFQHFSKLTNLRFLSLSWNTQLSLNFESN 299
Score = 50.1 bits (118), Expect = 2e-06
Identities = 30/77 (38%), Positives = 43/77 (54%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N L G IPKS G+L L SLDLS N L+ +P L NL N L+ + L++N
Sbjct: 1276 LNLSHNRLTGHIPKSIGNLTYLESLDLSLNMLTGVIPAELTNL------NFLEVMNLSNN 1437
Query: 381 QIIGTVPDMSGFSSLEN 397
++G +P F++ N
Sbjct: 1438 HLVGEIPRGKQFNTFTN 1488
Score = 41.2 bits (95), Expect = 0.001
Identities = 49/179 (27%), Positives = 75/179 (41%), Gaps = 20/179 (11%)
Frame = +3
Query: 333 PKSFGSLCSLRSLDLSSNKLSEDLPVMLH--------NLSVGC-----------AKNSLK 373
PK G +L LDLS+NKL +P L+ NLS N L
Sbjct: 363 PKLQGKFPNLDYLDLSNNKLDGRMPNWLYEKNSLKFLNLSQNYFMSIDQWINVNRSNGLS 542
Query: 374 ELYLASNQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGV 432
L L+ N + +P + SSLE + L N L G I + L L L N G
Sbjct: 543 GLDLSDNLLDDEIPLVVCNISSLEFLNLGYNNLTGIIPQCLAESTSLQVLNLQMNRFHGT 722
Query: 433 ITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPKWIRSQKN 491
+ S+F S + L+L N L +F ++ +L + L + +FP W+++ ++
Sbjct: 723 L-PSNFSKHSKIVSLNLYGNELEGRFPKSLFRCKKLEFLNLGVNKIEDNFPDWLQTMQD 896
Score = 37.4 bits (85), Expect = 0.013
Identities = 23/83 (27%), Positives = 41/83 (48%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
SL + N LEG PKS L L+L NK+ ++ P L + LK L L
Sbjct: 759 SLNLYGNELEGRFPKSLFRCKKLEFLNLGVNKIEDNFPDWLQTM------QDLKVLVLRD 920
Query: 380 NQIIGTVPDMSGFSSLENMFLYE 402
N++ G++ ++ S +++ +++
Sbjct: 921 NKLHGSLVNLKIKHSFQSLIIFD 989
>TC81387 weakly similar to PIR|T45645|T45645 receptor kinase-like protein -
Arabidopsis thaliana, partial (9%)
Length = 820
Score = 61.2 bits (147), Expect = 9e-10
Identities = 46/135 (34%), Positives = 70/135 (51%), Gaps = 1/135 (0%)
Frame = +2
Query: 319 HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLA 378
HSLI+++ SL G + + G+L L LDL +N LP L+ L LK L+++
Sbjct: 227 HSLILRNMSLRGTVSPNLGNLSFLVILDLKNNSFGGQLPTELYRL------RRLKILHIS 388
Query: 379 SNQIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
N+ G +P G S LE ++L N +G I ++ ++L L + N + G I +
Sbjct: 389 YNEFEGGIPAALGDLSQLEYLYLGVNNFSGFIPQSIGNLHQLKELGIGRNKMSGPIPQTI 568
Query: 438 FGNMSMLKYLSLSSN 452
NMS L+ L LSSN
Sbjct: 569 L-NMSSLEVLHLSSN 610
Score = 30.8 bits (68), Expect = 1.2
Identities = 21/57 (36%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Frame = +2
Query: 9 IEKERHALLELKSGLVLDDTY--LLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNS 63
I ++ ALL KS L+ D Y L +W T S C W G+ C + G V L L +
Sbjct: 83 ITTDQSALLAFKS-LITSDPYDVLANNWSTSSS-VCNWIGVTCDERHGRVHSLILRN 247
>CB893529 weakly similar to GP|8778255|gb| F12K21.25 {Arabidopsis thaliana},
partial (11%)
Length = 857
Score = 60.5 bits (145), Expect = 1e-09
Identities = 41/114 (35%), Positives = 57/114 (49%), Gaps = 1/114 (0%)
Frame = -1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L ++ N+L G IP S G+L L + L NKLS D+P M NL + L L+SN
Sbjct: 692 LDLQGNNLNGSIPSSIGNLGKLMEVQLGENKLSGDIPKMPLNLQIA--------LNLSSN 537
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
Q G +P + +LE + L N +G I + T L L L +N L GV+
Sbjct: 536 QFSGAIPSSFADLVNLEILDLSNNSFSGEIPPSLTKMVALTQLQLSNNHLSGVL 375
>TC82638 weakly similar to PIR|T00712|T00712 protein kinase homolog F22O13.7
- Arabidopsis thaliana, partial (27%)
Length = 1074
Score = 60.5 bits (145), Expect = 1e-09
Identities = 44/136 (32%), Positives = 69/136 (50%), Gaps = 1/136 (0%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
+II N EGGIPK FG+L L+ LDL+ + ++P L L + L ++L N
Sbjct: 659 MIIGYNEFEGGIPKEFGNLTKLKYLDLAEGNVGGEIPDELGKLKL------LNTVFLYKN 820
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
G +P ++ +SL + L +N+L+G I + L L N L G + S G
Sbjct: 821 SFEGKIPTNIGNMTSLVLLDLSDNMLSGNIPAEISQLKNLQLLNFMRNKLSGPV-PSGLG 997
Query: 440 NMSMLKYLSLSSNSLA 455
++ L+ L L +NSL+
Sbjct: 998 DLPQLEVLELWNNSLS 1045
Score = 33.5 bits (75), Expect = 0.19
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 2/63 (3%)
Frame = +1
Query: 2 LLVSGSCIEKERHALLELKSGLVLDDTYLLPSWDT--KSDDCCAWEGIGCRNQTGHVEIL 59
L +G+ + E ALL +K+GL+ D L W + C W G+ C N G VE L
Sbjct: 52 LCCAGAAADNEAFALLSIKAGLI-DPLNSLHDWKDGGAAQAHCNWTGVQC-NSAGAVEKL 225
Query: 60 DLN 62
+L+
Sbjct: 226 NLS 234
Score = 32.3 bits (72), Expect = 0.42
Identities = 18/45 (40%), Positives = 22/45 (48%)
Frame = +1
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS 364
SL + N GG P G L +L+ SSN S LP L N+S
Sbjct: 364 SLDVSQNFFTGGFPLGLGKASELLTLNASSNNFSGFLPEDLGNIS 498
>BM779095 weakly similar to PIR|B84431|B8 probable receptor protein kinase
[imported] - Arabidopsis thaliana, partial (15%)
Length = 721
Score = 60.1 bits (144), Expect = 2e-09
Identities = 44/139 (31%), Positives = 72/139 (51%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
SL + SN + GGIPK+ G L L+SLDLS N+++ +P L N+ SL EL L+
Sbjct: 234 SLNLASNFISGGIPKALGQLNKLQSLDLSHNQITGWIPSELSNVC-----GSLLELKLSF 398
Query: 380 NQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
N I G+VP GF+S + L + + +N++ G + +S
Sbjct: 399 NNITGSVP--FGFNSCTWLQLVD---------------------ISNNNMTGELPESVIR 509
Query: 440 NMSMLKYLSLSSNSLALKF 458
++ L+ L L +N++++KF
Sbjct: 510 SLGSLQELRLGNNAISMKF 566
Score = 56.2 bits (134), Expect = 3e-08
Identities = 53/174 (30%), Positives = 81/174 (46%), Gaps = 3/174 (1%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
SL + N+L G I SL LDLS N LS+ P+ L N + SLK L LAS
Sbjct: 90 SLDLSFNNLTGSISDIKIDCKSLLQLDLSGNNLSDSFPISLSNCT------SLKSLNLAS 251
Query: 380 NQIIGTVPDMSG-FSSLENMFLYENLLNGTILKN-STFPYRLANLYLDSNDLDGVITDSH 437
N I G +P G + L+++ L N + G I S L L L N++ G +
Sbjct: 252 NFISGGIPKALGQLNKLQSLDLSHNQITGWIPSELSNVCGSLLELKLSFNNITGSVPFG- 428
Query: 438 FGNMSMLKYLSLSSNSLALKFSENWVPPF-QLSTIYLRSCTLGPSFPKWIRSQK 490
F + + L+ + +S+N++ + E+ + L + L + + FP I S K
Sbjct: 429 FNSCTWLQLVDISNNNMTGELPESVIRSLGSLQELRLGNNAISMKFPSSISSLK 590
>BG453331 weakly similar to GP|20521202|dbj putative Cf2/Cf5 disease
resistance protein {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 589
Score = 59.3 bits (142), Expect = 3e-09
Identities = 32/69 (46%), Positives = 44/69 (63%)
Frame = +2
Query: 5 SGSCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSD 64
S C+E+ER ALLE+K G D + L SW K +DCC W+GI C N TGHV +DL +
Sbjct: 122 SFGCMEQERKALLEIK-GSFNDPLFRLSSW--KGNDCCKWKGISCSNITGHVVKIDLRNP 292
Query: 65 QFGPFEGDR 73
+ P +G++
Sbjct: 293 CY-PQKGEQ 316
>BF648743 weakly similar to PIR|H84632|H846 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (9%)
Length = 648
Score = 58.9 bits (141), Expect = 4e-09
Identities = 54/175 (30%), Positives = 83/175 (46%), Gaps = 25/175 (14%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS----VGCAKN------ 370
L I+ ++L G IP+ G L +L +DLS N LS +P + NLS + + N
Sbjct: 97 LAIQKSNLVGSIPQEIGFLTNLAYIDLSKNSLSGGIPETIGNLSKLDTLVLSNNTKMSGP 276
Query: 371 ---------SLKELYLASNQIIGTVPD-MSGFSSLENMFLYENLLNGTILKNSTFPYRLA 420
SL LY + + G++PD + +L+ + L N L+G+I L
Sbjct: 277 IPHSLWNMSSLTVLYFDNIGLSGSIPDSIQNLVNLKELALDINHLSGSIPSTIGDLKNLI 456
Query: 421 NLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSL-----ALKFSENWVPPFQLST 470
LYL SN+L G I S GN+ L+ LS+ N+L A + W+ F++ T
Sbjct: 457 KLYLGSNNLSGPIPAS-IGNLINLQVLSVQENNLTGTIPASIGNLKWLTVFEVXT 618
>TC86726 similar to GP|14626935|gb|AAK70805.1 leucine-rich repeat resistance
protein-like protein {Gossypium hirsutum}, partial (92%)
Length = 1507
Score = 58.9 bits (141), Expect = 4e-09
Identities = 44/134 (32%), Positives = 66/134 (48%), Gaps = 1/134 (0%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N L G IP G+L +LR LD +N L + ++ GC SL+ LYL +N
Sbjct: 722 LYLHENRLTGRIPPELGTLQNLRHLDAGNNHLVGTIRELIR--IEGCFP-SLRNLYLNNN 892
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
G +P ++ SSLE ++L N ++G I + +L LYLD N G I + F
Sbjct: 893 YFTGGIPAQLANLSSLEILYLSYNKMSGVIPSSVAHIPKLTYLYLDHNQFSGRIPEP-FY 1069
Query: 440 NMSMLKYLSLSSNS 453
LK + + N+
Sbjct: 1070KHPFLKEMYIEGNA 1111
Score = 45.4 bits (106), Expect = 5e-05
Identities = 39/129 (30%), Positives = 58/129 (44%), Gaps = 1/129 (0%)
Frame = +2
Query: 327 SLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTV 386
S+ G P + SL L LDL +NKL+ +P + L LK L L N++ +
Sbjct: 452 SIVGPFPTAVTSLLDLTRLDLHNNKLTGPIPPQIGRL------KRLKILNLRWNKLQDAI 613
Query: 387 -PDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLK 445
P++ SL +++L N G I + L LYL N L G I G + L+
Sbjct: 614 PPEIGELKSLTHLYLSFNSFKGEIPRELADLPDLRYLYLHENRLTGRI-PPELGTLQNLR 790
Query: 446 YLSLSSNSL 454
+L +N L
Sbjct: 791 HLDAGNNHL 817
Score = 40.4 bits (93), Expect = 0.002
Identities = 30/100 (30%), Positives = 45/100 (45%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + +N GGIP +L SL L LS NK+S +P + ++ L LYL
Sbjct: 872 NLYLNNNYFTGGIPAQLANLSSLEILYLSYNKMSGVIPSSVAHIP------KLTYLYLDH 1033
Query: 380 NQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRL 419
NQ G +P+ ++ FL E + G + P L
Sbjct: 1034NQFSGRIPE----PFYKHPFLKEMYIEGNAFRPGVNPIGL 1141
>TC80900 weakly similar to PIR|G84652|G84652 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana, partial (24%)
Length = 1054
Score = 58.9 bits (141), Expect = 4e-09
Identities = 52/164 (31%), Positives = 79/164 (47%), Gaps = 1/164 (0%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N L G IPKS +L +L SLDLS N LS ++ ++ NL L+ L+L SN
Sbjct: 173 LFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNL------QKLEILHLFSN 334
Query: 381 QIIGTVPD-MSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
G +P+ ++ L+ + L+ N L G I + L L L SN+L G I +S
Sbjct: 335 NFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCA 514
Query: 440 NMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFP 483
+ ++ K + L SNSL + + L + L+ L P
Sbjct: 515 SKNLHKII-LFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLP 643
Score = 52.8 bits (125), Expect = 3e-07
Identities = 50/167 (29%), Positives = 75/167 (43%), Gaps = 1/167 (0%)
Frame = +2
Query: 319 HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLA 378
H +I+ SNSL+G IPK S +L + L N LS LP+ + L + L ++
Sbjct: 527 HKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLP------QIYLLDIS 688
Query: 379 SNQIIGTVPDMS-GFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
N+ G + D SL+ + L N +G L NS ++ L L N G I
Sbjct: 689 GNKFSGKINDRKWNMPSLQMLNLANNNFSGD-LPNSFGGNKVEGLDLSQNQFSGYI-QIG 862
Query: 438 FGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFPK 484
F N+ L L L++N+L KF E +L ++ L L P+
Sbjct: 863 FKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPE 1003
>BG644528 weakly similar to GP|21391894|g systemin receptor SR160
{Lycopersicon peruvianum}, partial (10%)
Length = 762
Score = 57.4 bits (137), Expect = 1e-08
Identities = 47/134 (35%), Positives = 63/134 (46%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L I +N G I S G L LDL NKL+ +P+ + LS L LYL N
Sbjct: 365 LAIGNNQFSGRIHASIGRCKRLSFLDLRMNKLAGVIPMEIFQLS------GLTTLYLHGN 526
Query: 381 QIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGN 440
+ G++P LE M + +N L+G I K L L + N+ G I +S G+
Sbjct: 527 SLNGSLPPQFKMEQLEAMVVSDNKLSGNIPKIEV--NGLKTLMMARNNFSGSIPNS-LGD 697
Query: 441 MSMLKYLSLSSNSL 454
+ L L LSSNSL
Sbjct: 698 LPSLVTLDLSSNSL 739
Score = 37.4 bits (85), Expect = 0.013
Identities = 18/38 (47%), Positives = 25/38 (65%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP 357
+L++ N+ G IP S G L SL +LDLSSN L+ +P
Sbjct: 641 TLMMARNNFSGSIPNSLGDLPSLVTLDLSSNSLTGPIP 754
>BE320710 weakly similar to GP|16924044|gb| Putative protein kinase {Oryza
sativa}, partial (12%)
Length = 476
Score = 57.4 bits (137), Expect = 1e-08
Identities = 40/115 (34%), Positives = 59/115 (50%), Gaps = 1/115 (0%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + N+L G IPK G L L L+LS NK ++PV L+V ++ L L+
Sbjct: 84 ALELAINNLSGFIPKKLGMLSMLLQLNLSQNKFEGNIPVEFGQLNV------IENLDLSG 245
Query: 380 NQIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
N + GT+P M G + LE + L N L+GTI + L + + N L+G I
Sbjct: 246 NSMNGTIPAMLGQLNHLETLNLSHNNLSGTIPSSFVDMLSLTTVDISYNQLEGPI 410
>TC77085 similar to GP|21536600|gb|AAM60932.1 putative disease resistance
protein {Arabidopsis thaliana}, partial (88%)
Length = 1662
Score = 57.4 bits (137), Expect = 1e-08
Identities = 43/141 (30%), Positives = 67/141 (47%), Gaps = 1/141 (0%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
+ I++N+L G IP++ GS+ L + L NK + +P + L+ L +L L +N
Sbjct: 440 IYIENNTLSGPIPQNIGSMNQLEAFSLQENKFTGPIPSSISALT------KLTQLKLGNN 601
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
+ GT+P + ++L + L N L+G I T L L L N G I S
Sbjct: 602 FLTGTIPVSLKNLTNLTYLSLQGNQLSGNIPDIFTSLKNLIILQLSHNKFSGNIPLSISS 781
Query: 440 NMSMLKYLSLSSNSLALKFSE 460
L+YL L NSL+ K +
Sbjct: 782 LYPTLRYLELGHNSLSGKIPD 844
Score = 48.1 bits (113), Expect = 7e-06
Identities = 51/176 (28%), Positives = 78/176 (43%), Gaps = 19/176 (10%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVM----LHNLSVGCAKNSLKEL 375
+L + N +G +PKSF +L + +LDLS N L + PVM + +L + LKE+
Sbjct: 872 TLDLSKNQFKGTVPKSFANLTKIFNLDLSDNFLVDPFPVMNVKGIESLDLSRNMFHLKEI 1051
Query: 376 --YLASNQII--------GTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYL- 424
++A++ II G + + LE F L+G + S YL
Sbjct: 1052 PKWVATSPIIYSLKLAHCGIKMKLDDWKPLETFFYDYIDLSGNEISGSAVGLLNKTEYLI 1231
Query: 425 ----DSNDLDGVITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSC 476
N L + FGN LKYL LS N + K +++ V +L+ Y R C
Sbjct: 1232 EFRGSENLLKFDLESLKFGN--RLKYLDLSHNLVFGKVTKSVVGIQKLNVSYNRLC 1393
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/61 (36%), Positives = 28/61 (45%)
Frame = +2
Query: 8 CIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSDQFG 67
C + LL KSG+ D T +L SW +CC W G+GC + V L L D
Sbjct: 128 CDPDDESGLLAFKSGIKSDPTSMLKSW-IPGTNCCTWVGVGCLDNK-RVTSLSLTGDTEN 301
Query: 68 P 68
P
Sbjct: 302 P 304
Score = 35.0 bits (79), Expect = 0.065
Identities = 35/135 (25%), Positives = 57/135 (41%), Gaps = 5/135 (3%)
Frame = +2
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
NSL G IP G +L +LDLS N+ +P NL+ + L L+ N ++
Sbjct: 818 NSLSGKIPDFLGKFKALDTLDLSKNQFKGTVPKSFANLT------KIFNLDLSDNFLVDP 979
Query: 386 VPDMS--GFSSLE---NMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGN 440
P M+ G SL+ NMF + + + +LA+ + D ++ F
Sbjct: 980 FPVMNVKGIESLDLSRNMFHLKEIPKWVATSPIIYSLKLAHCGIKMKLDDWKPLETFF-- 1153
Query: 441 MSMLKYLSLSSNSLA 455
Y+ LS N ++
Sbjct: 1154---YDYIDLSGNEIS 1189
>BI308178 weakly similar to GP|6714444|gb|A putative disease resistance
protein {Arabidopsis thaliana}, partial (7%)
Length = 602
Score = 57.4 bits (137), Expect = 1e-08
Identities = 26/59 (44%), Positives = 36/59 (60%)
Frame = +2
Query: 5 SGSCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNS 63
+ C+EKE+ ALL+ + + L + SW K ++CC WEGI C N TGHV LDL +
Sbjct: 83 TNKCVEKEKRALLKFRDAINLKYRDGISSW--KGEECCKWEGISCDNLTGHVTSLDLEA 253
Score = 40.4 bits (93), Expect = 0.002
Identities = 30/79 (37%), Positives = 38/79 (47%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
SL + N EG IPK GSL L L+L N L +P L NLS +L+ L L
Sbjct: 320 SLNLHGNQFEGKIPKCIGSLDKLIELNLHDNNLVSVIPPSLGNLS------NLQTLDLGY 481
Query: 380 NQIIGTVPDMSGFSSLENM 398
N + T D+ S L N+
Sbjct: 482 NSL--TTNDLEWLSHLSNL 532
Score = 28.5 bits (62), Expect = 6.1
Identities = 23/60 (38%), Positives = 30/60 (49%)
Frame = +2
Query: 395 LENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSL 454
L ++ L+ N G I K +L L L N+L VI S GN+S L+ L L NSL
Sbjct: 314 LTSLNLHGNQFEGKIPKCIGSLDKLIELNLHDNNLVSVIPPS-LGNLSNLQTLDLGYNSL 490
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.146 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,274,520
Number of Sequences: 36976
Number of extensions: 274622
Number of successful extensions: 2736
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 2505
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2671
length of query: 491
length of database: 9,014,727
effective HSP length: 100
effective length of query: 391
effective length of database: 5,317,127
effective search space: 2078996657
effective search space used: 2078996657
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136507.3


