

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
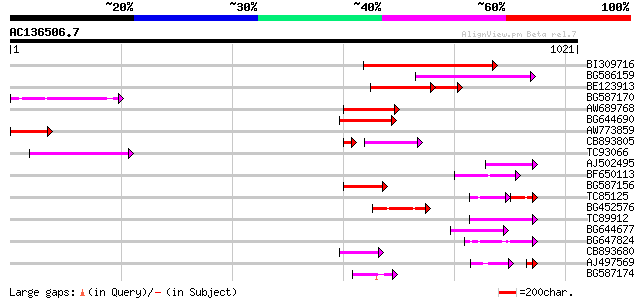
Query= AC136506.7 - phase: 0 /pseudo
(1021 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 277 2e-74
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 155 6e-38
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 102 3e-32
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 121 2e-27
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 97 3e-20
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 92 8e-19
AW773859 91 2e-18
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 75 1e-17
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 79 7e-15
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 75 2e-13
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 74 2e-13
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 72 8e-13
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 51 1e-12
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 67 3e-11
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 65 1e-10
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 65 1e-10
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 60 4e-09
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 55 2e-07
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 40 6e-07
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 47 3e-05
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 277 bits (708), Expect = 2e-74
Identities = 138/241 (57%), Positives = 177/241 (73%)
Frame = +2
Query: 637 QVCKLSKSLYGLKQASRKWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDI 696
+VC+L KS+YGLKQASR+WY KL+ LIS GY Q++SD SLFTK S T LLVYVDDI
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 697 ILAGDSLIEIAFIKNVINQAFKIKDLGTLKYFLGLEVAHSQSGISLCQRKYCLDLLHDLG 756
+LAG+ + EI +K + FKIKDLG+L+YFLGLEVA S+ GI L QRKY L+LL D G
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 757 LLGSKPVTTPSDPSIKLHNDSSPLFADVSAYRRLVGRLIYLNTTRPDITFITQQLSQFLS 816
L K TP D S+KLHN SPL+ D + YRRL+G+LIYL TTRPDI+F QQLSQF+S
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 817 KPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSLHIQGYTNADWAGCKDTRRSISCRCFFL 876
KP HY AA+RV++YLK +P +G+F+ +S+L + + ++DWA C TR+S++ FL
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 877 G 877
G
Sbjct: 737 G 739
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 155 bits (393), Expect = 6e-38
Identities = 77/217 (35%), Positives = 124/217 (56%)
Frame = +1
Query: 731 LEVAHSQSGISLCQRKYCLDLLHDLGLLGSKPVTTPSDPSIKLHNDSSPLFADVSAYRRL 790
+EV ++ GI +CQRKY DLL G+ S P P KL D + + D + Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 791 VGRLIYLNTTRPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSLH 850
VG L+YL TRPD+ ++ +S+F++ PT H +A RV++YL G+ GI + R+ S
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 851 IQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCEL 910
++ YT++D+AG D R+S S F L +SW +KK P ++ +++AE+ A A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 911 QWLLYLLRDLHIRCVKLLVLYCDNQSVMHIAANPDFH 947
W+ +L L + +YCDN S + ++ NP H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 102 bits (254), Expect(2) = 3e-32
Identities = 54/118 (45%), Positives = 77/118 (64%), Gaps = 1/118 (0%)
Frame = +1
Query: 650 QASRKWYEKLTGLLISNGYQQATSDASLFTKKAS-VSLTILLVYVDDIILAGDSLIEIAF 708
Q+ R W+++ T ++ GY Q +D ++F K +S V IL+VYVDDI L GD I
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 709 IKNVINQAFKIKDLGTLKYFLGLEVAHSQSGISLCQRKYCLDLLHDLGLLGSKPVTTP 766
+KN++ + F+IKDLG LKYFLG+EVA + G S+ QRKY LDLL + ++G K + P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 55.8 bits (133), Expect(2) = 3e-32
Identities = 27/58 (46%), Positives = 37/58 (63%)
Frame = +2
Query: 758 LGSKPVTTPSDPSIKLHNDSSPLFADVSAYRRLVGRLIYLNTTRPDITFITQQLSQFL 815
L KP TP D ++KL + D Y+RLVG+LIYL+ TRPDI+F+ +SQF+
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 121 bits (303), Expect = 2e-27
Identities = 71/207 (34%), Positives = 110/207 (52%), Gaps = 3/207 (1%)
Frame = -3
Query: 1 IWHFRLGHLSNQRLSKMHQLYPSISVDNKATCDICPFAKQRKLPYNFSHSIAKSKFELLY 60
+WH RLGH + L+ M P + +NK C+ C K K + + ++ ++ F+L+Y
Sbjct: 584 LWHARLGHPHGRALNLM---LPGVVFENK-NCEACILGKHCKNVFPRTSTVYENCFDLIY 417
Query: 61 FDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPK 120
D+W S HKYF+T +D+ S++ W+ L+ +K V K F + H+H K
Sbjct: 416 TDLW-TAPSLSRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIK 240
Query: 121 FIRSDNGPEFLLLEFYA---SKGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKL 177
+RSDNG E+ F + GI+HQ SC TP+QN +RK++H++ VAR+L FQ
Sbjct: 239 ILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN- 63
Query: 178 PNIFWSYAVLYSVFLINRVPTPFLHHK 204
V + +LIN +PT L K
Sbjct: 62 --------VSTACYLINWIPTKVLRIK 6
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 97.1 bits (240), Expect = 3e-20
Identities = 47/101 (46%), Positives = 64/101 (62%)
Frame = +1
Query: 602 WFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASRKWYEKLTG 661
W + Q+D+NNAFL+G L E+VYM+ G + + VCKL+KSLYGLKQA R WYE LT
Sbjct: 367 WEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWYEXLTS 546
Query: 662 LLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDS 702
I G+ ++ D SL + + L +YVDDI++ G S
Sbjct: 547 AQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGSS 669
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 92.4 bits (228), Expect = 8e-19
Identities = 47/104 (45%), Positives = 70/104 (67%), Gaps = 1/104 (0%)
Frame = -2
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSK-PNQVCKLSKSLYGLKQAS 652
I A+ + L+Q+DV +AF++GDL E+V++ PG ++ PN V +L+K+LYGLKQA
Sbjct: 313 IAFAAFMGFKLYQMDVKSAFINGDLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAP 134
Query: 653 RKWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDI 696
R WYE+L+ L+ NG+++ D +LF K L I+ VYVDDI
Sbjct: 133 RAWYERLSKFLLKNGFKRGKIDNTLFLLKRE*ELLIIQVYVDDI 2
>AW773859
Length = 538
Score = 90.9 bits (224), Expect = 2e-18
Identities = 35/76 (46%), Positives = 53/76 (69%)
Frame = -3
Query: 1 IWHFRLGHLSNQRLSKMHQLYPSISVDNKATCDICPFAKQRKLPYNFSHSIAKSKFELLY 60
+WHFRLGHLSN++L +H +P I++D + CDIC +++ +KLP+ S + A +EL +
Sbjct: 230 LWHFRLGHLSNRKLLSLHSNFPFITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYELFH 51
Query: 61 FDIWGPLAQTSIHGHK 76
FDIWGP + SIH +
Sbjct: 50 FDIWGPFSTQSIHNQR 3
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 75.1 bits (183), Expect(2) = 1e-17
Identities = 35/105 (33%), Positives = 60/105 (56%), Gaps = 1/105 (0%)
Frame = +3
Query: 640 KLSKSLYGLKQASRKWYEKLTGLLISNGYQQATSDASLFTKKASVS-LTILLVYVDDIIL 698
++ ++LYGLKQA R WY ++ G+++ + +LF K + + I+ +YVDD+I
Sbjct: 462 RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIF 641
Query: 699 AGDSLIEIAFIKNVINQAFKIKDLGTLKYFLGLEVAHSQSGISLC 743
G+ K + + F + DLG + YFLG+EV ++ GI +C
Sbjct: 642 IGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYIC 776
Score = 33.9 bits (76), Expect(2) = 1e-17
Identities = 12/23 (52%), Positives = 21/23 (91%)
Frame = +2
Query: 602 WFLHQLDVNNAFLHGDLHEDVYM 624
W ++QLDV +AFL+G+L+E+V++
Sbjct: 353 WSVYQLDVKSAFLYGELNEEVFV 421
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 79.3 bits (194), Expect = 7e-15
Identities = 56/191 (29%), Positives = 88/191 (45%), Gaps = 5/191 (2%)
Frame = +1
Query: 37 FAKQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKN 96
F ++K+ ++ + K + ++ D+WGP TS G +Y +TI+DDF R +WV L+
Sbjct: 40 FGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 97 KAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEFL---LLEFYASKGIIHQKSCVETPEQ 153
K E K + +++T K + +DN EF EF + GI K+ P+Q
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 154 NARVERKHQHILNVARALPFQFKLPN--IFWSYAVLYSVFLINRVPTPFLHHKSPY*VLY 211
N ER + +L AR + L N W A + L+NR P L K P +
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWS 579
Query: 212 DSLPDIQFFKI 222
+L D +I
Sbjct: 580 GNLVDYSNLRI 612
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 74.7 bits (182), Expect = 2e-13
Identities = 37/93 (39%), Positives = 53/93 (56%)
Frame = +2
Query: 857 ADWAGCKDTRRSISCRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYL 916
+DWAG +TR+S S F LG ISW +KK P ++ ++EAEY A S + WL +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 917 LRDLHIRCVKLLVLYCDNQSVMHIAANPDFHSR 949
L +H +YCDN+S + ++ NP FH R
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGR 280
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 74.3 bits (181), Expect = 2e-13
Identities = 44/123 (35%), Positives = 67/123 (53%), Gaps = 3/123 (2%)
Frame = +1
Query: 801 RPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFP---RSSSLHIQGYTNA 857
RPDI + +S+F+ P H AA R+++Y++G+ G+ FP +S + Y+++
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 858 DWAGCKDTRRSISCRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLL 917
DW G RRS S F + ISW TKK P + S EAEY A AT + WL ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 918 RDL 920
++L
Sbjct: 472 KEL 480
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 72.4 bits (176), Expect = 8e-13
Identities = 36/79 (45%), Positives = 51/79 (63%), Gaps = 1/79 (1%)
Frame = -1
Query: 602 WFLHQLDVNNAFLHGDLHEDVYMTVLPGVT-TSKPNQVCKLSKSLYGLKQASRKWYEKLT 660
W L Q+DV NAFL G+L ++VYM PG+ K V +L K++YGLKQ+ R WY KL+
Sbjct: 252 WGLWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLS 73
Query: 661 GLLISNGYQQATSDASLFT 679
L G++++ D +LFT
Sbjct: 72 TTLNGRGFRKSELDHTLFT 16
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 51.2 bits (121), Expect(2) = 1e-12
Identities = 26/75 (34%), Positives = 45/75 (59%)
Frame = +1
Query: 828 RVIKYLKGSPGRGIFFPRSSSLHIQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTKK 887
R+++Y+KG+ G + F S L ++GY ++D+AG D R+S + F L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 888 *PTISRFSSEAEYRA 902
++ ++EAEY A
Sbjct: 184 QTVVALSTTEAEYMA 228
Score = 40.4 bits (93), Expect(2) = 1e-12
Identities = 20/48 (41%), Positives = 30/48 (61%)
Frame = +3
Query: 902 AMASATCELQWLLYLLRDLHIRCVKLLVLYCDNQSVMHIAANPDFHSR 949
++ A E W+ L+ +L + ++ V YCD+QS +HIA NP FHSR
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITV-YCDSQSALHIARNPAFHSR 368
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 67.4 bits (163), Expect = 3e-11
Identities = 45/106 (42%), Positives = 65/106 (60%)
Frame = +2
Query: 653 RKWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNV 712
RK Y KL LIS GY+Q+ +D S+F+ TI LVY+DDI+ A D E +K+
Sbjct: 236 RK*Y*KLLSTLISLGYKQSPNDHSIFS--FGRRFTIFLVYLDDIVFARDDHSETQLVKSH 409
Query: 713 INQAFKIKDLGTLKYFLGLEVAHSQSGISLCQRKYCLDLLHDLGLL 758
+++ F I LGTL Y +G+++A S+S I Q KY ++LL + G L
Sbjct: 410 LDKNFIIIGLGTLHY-VGVKIA*SES*IIDDQCKYTIELLEESGQL 544
Score = 30.4 bits (67), Expect = 3.6
Identities = 41/152 (26%), Positives = 67/152 (43%), Gaps = 16/152 (10%)
Frame = +3
Query: 638 VCKLSKSLYGLKQASRKWYEK-LTGLLISNGYQQ----ATSDASLFTKKASVSLTILLVY 692
VCK S+YGLK A R+ Y K G I + YQ ++ + T +++ +L +
Sbjct: 165 VCKFHNSIYGLKHAHRQ*YSKFFEGNDIESCYQL*FL*DINNLQMITPFLVLAVGLLFFW 344
Query: 693 VDDIILAGDSLIEIAFIKNVINQAFKIKDLGTLKYFLGLEVAHS-QSGISLCQRKYCLDL 751
+IL L+ I N+++ + ++ AHS SG L ++ L +
Sbjct: 345 YIWMILF--LLVTIILKLNLLSPIW-------IRIS*S*V*AHSIMSGSKLLNQRVKLLM 497
Query: 752 LHD----------LGLLGSKPVTTPSDPSIKL 773
+ + LG KP +TP DPS+KL
Sbjct: 498 ISANILLSCLKKVVS*LGMKPSSTPRDPSLKL 593
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 65.5 bits (158), Expect = 1e-10
Identities = 37/123 (30%), Positives = 70/123 (56%), Gaps = 2/123 (1%)
Frame = +1
Query: 829 VIKYLKGSPGRGIFFPRSSSLH--IQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTK 886
V+KYL S + + +++ ++GY +AD+AG DTR+S+S F L + ISW+
Sbjct: 16 VLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKAN 195
Query: 887 K*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLYCDNQSVMHIAANPDF 946
+ ++ +++AEY A + WL ++ +L I + + ++CD+QS +H+A + +
Sbjct: 196 QQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSAIHLANHQVY 372
Query: 947 HSR 949
H R
Sbjct: 373 HER 381
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 65.1 bits (157), Expect = 1e-10
Identities = 39/104 (37%), Positives = 54/104 (51%)
Frame = -3
Query: 795 IYLNTTRPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSLHIQGY 854
I L P+ITF LS++ S PT H N + KYLKG G+F+ + S + GY
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 855 TNADWAGCKDTRRSISCRCFFLGQSLISWRTKK*PTISRFSSEA 898
NA + RS + F G ++ISWR+ K* TI+ S+ A
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 60.1 bits (144), Expect = 4e-09
Identities = 49/135 (36%), Positives = 70/135 (51%), Gaps = 4/135 (2%)
Frame = -3
Query: 819 THTHYNAALRVIKYLKGSPGRGIFFPRSSSLHIQGYTNAD---WAGCKDTRRSISCRCFF 875
T H AA++++ YLK SP + IFFP S + I+ + ++D A +S+
Sbjct: 599 TAQHPQAAIQIL-YLKISPSQ*IFFP--S*IQIKAFCDSD*IDQAA*TLENQSVIFASS* 429
Query: 876 LGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLYCDNQ 935
S KK R++ + YR++ S CE++WL YLL DL +K +LYCDNQ
Sbjct: 428 ATHSYAGNLKKK-----RYNFKI-YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQ 267
Query: 936 S-VMHIAANPDFHSR 949
S HIAAN F R
Sbjct: 266 SAARHIAANSSFLER 222
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 54.7 bits (130), Expect = 2e-07
Identities = 31/80 (38%), Positives = 42/80 (51%)
Frame = -2
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ + +I + +L LDV AFL GDL ED+YM G + V KL KS+YGLKQ R
Sbjct: 515 LSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGKLKKSMYGLKQGPR 336
Query: 654 KWYEKLTGLLISNGYQQATS 673
+ L L G++ S
Sbjct: 335 QCI*SLKALCTRKGFRSEIS 276
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 40.4 bits (93), Expect(2) = 6e-07
Identities = 31/78 (39%), Positives = 43/78 (54%)
Frame = +3
Query: 830 IKYLKGSPGRGIFFPRSSSLHIQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTKK*P 889
+ YLK +PG+ IF +S+ H C + R + CF L SLISW++KK
Sbjct: 15 LHYLK-TPGKCIFVSNASNPHFNR------GSCPYSIR*TTEFCF-LSSSLISWKSKKQC 170
Query: 890 TISRFSSEAEYRAMASAT 907
+SR SEA RA+A+AT
Sbjct: 171 VVSRSFSEA**RALANAT 224
Score = 32.0 bits (71), Expect(2) = 6e-07
Identities = 13/19 (68%), Positives = 14/19 (73%)
Frame = +2
Query: 931 YCDNQSVMHIAANPDFHSR 949
YCDN S +HIAAN FH R
Sbjct: 290 YCDNISALHIAANMVFHER 346
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 47.4 bits (111), Expect = 3e-05
Identities = 32/91 (35%), Positives = 48/91 (52%), Gaps = 10/91 (10%)
Frame = -1
Query: 617 DLHEDVYMTVLPG-VTTSKPNQVCKLSKSLYGLKQASRKWYEKL---------TGLLISN 666
+L E +YMT G + K + VCKL KSLYGLKQ+ R+WY++ TG+L+
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRSSWATTGVLM-- 76
Query: 667 GYQQATSDASLFTKKASVSLTILLVYVDDII 697
+ S T+ L++YVDD++
Sbjct: 75 ------TVVST*TR*RMSRYIYLVLYVDDML 1
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.340 0.149 0.489
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,876,159
Number of Sequences: 36976
Number of extensions: 676345
Number of successful extensions: 9705
Number of sequences better than 10.0: 241
Number of HSP's better than 10.0 without gapping: 5434
Number of HSP's successfully gapped in prelim test: 696
Number of HSP's that attempted gapping in prelim test: 3078
Number of HSP's gapped (non-prelim): 7103
length of query: 1021
length of database: 9,014,727
effective HSP length: 106
effective length of query: 915
effective length of database: 5,095,271
effective search space: 4662172965
effective search space used: 4662172965
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC136506.7


