

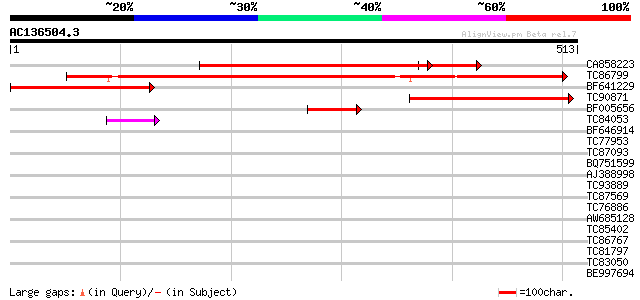
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.3 + phase: 0 /pseudo
(513 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA858223 similar to GP|22655368|gb At2g17760/At2g17760 {Arabidop... 398 e-111
TC86799 similar to GP|22655368|gb|AAM98276.1 At2g17760/At2g17760... 301 5e-82
BF641229 weakly similar to GP|22655368|gb| At2g17760/At2g17760 {... 287 8e-78
TC90871 similar to PIR|T45765|T45765 hypothetical protein F24M12... 276 1e-74
BF005656 93 3e-19
TC84053 similar to PIR|T04698|T04698 hypothetical protein F4B14.... 47 2e-05
BF646914 similar to GP|22655368|gb| At2g17760/At2g17760 {Arabido... 37 0.014
TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-bindin... 35 0.053
TC87093 homologue to GP|10177817|dbj|BAB11183. gene_id:MKD15.14~... 35 0.069
BQ751599 similar to GP|7293866|gb| ftz-f1 gene product {Drosophi... 33 0.34
AJ388998 similar to SP|Q9SS17|RS24_ 40S ribosomal protein S24. [... 32 0.45
TC93889 32 0.45
TC87569 similar to PIR|E96776|E96776 hypothetical protein F25A4.... 32 0.45
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 32 0.45
AW685128 homologue to GP|1556394|emb| GATA factor {Drosophila me... 32 0.76
TC85402 similar to GP|21554238|gb|AAM63313.1 Contains similarity... 32 0.76
TC86767 similar to PIR|T52470|T52470 cytochrome-b5 reductase (EC... 31 1.00
TC81797 similar to GP|17065224|gb|AAL32766.1 Unknown protein {Ar... 31 1.00
TC83050 similar to SP|Q9FGZ4|WR48_ARATH Probable WRKY transcript... 27 1.2
BE997694 similar to GP|13646986|db DNA-binding protein DF1 {Pisu... 31 1.3
>CA858223 similar to GP|22655368|gb At2g17760/At2g17760 {Arabidopsis
thaliana}, partial (32%)
Length = 813
Score = 398 bits (1023), Expect = e-111
Identities = 205/211 (97%), Positives = 207/211 (97%)
Frame = +1
Query: 172 TCVNKHNAIHPVAVVGMKLSIFQMILHLLDS*LKMCYI*LQIMIKQKILTHKLPLVVAKF 231
TCVNKHNAIHPVAVVGMKLSIFQMILHLLDS*LKMCYI*LQIMIKQKILTHKLPLVVAKF
Sbjct: 1 TCVNKHNAIHPVAVVGMKLSIFQMILHLLDS*LKMCYI*LQIMIKQKILTHKLPLVVAKF 180
Query: 232 RLVCF*MVQLQMVYLDLVWKMYLFRAS*PKRGLFQILFQCVLDLMVLEESRLGIPAAQTK 291
RLVCF*MVQLQMVYLDLVWKMYLFRAS*PKRGLFQILFQCVLDLMVLEESRLGIPAAQTK
Sbjct: 181 RLVCF*MVQLQMVYLDLVWKMYLFRAS*PKRGLFQILFQCVLDLMVLEESRLGIPAAQTK 360
Query: 292 EKHHSTSGNHILHITSPLHKLLWEDMLPIMSFMQFSTPVPHSHT*MTQLIR*FPRSSIL* 351
EKHHSTSGNHILHITSPLHKLLWEDMLPIMSFMQFSTPVPHSHT*MTQLIR*FPRSSIL*
Sbjct: 361 EKHHSTSGNHILHITSPLHKLLWEDMLPIMSFMQFSTPVPHSHT*MTQLIR*FPRSSIL* 540
Query: 352 SKQIDIRLCHLIRISPLNTVMT*VQIRQLKF 382
SKQIDIRLCHLIRISPLNTVMT* Q ++ F
Sbjct: 541 SKQIDIRLCHLIRISPLNTVMT*DQTIEVPF 633
Score = 97.4 bits (241), Expect = 1e-20
Identities = 52/57 (91%), Positives = 55/57 (96%)
Frame = +2
Query: 371 VMT*VQIRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVTI*ISLD 427
++ * +IRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVTI*ISLD
Sbjct: 593 ILL*HEIRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVTI*ISLD 763
>TC86799 similar to GP|22655368|gb|AAM98276.1 At2g17760/At2g17760
{Arabidopsis thaliana}, partial (48%)
Length = 1850
Score = 301 bits (770), Expect = 5e-82
Identities = 225/467 (48%), Positives = 289/467 (61%), Gaps = 14/467 (2%)
Frame = +2
Query: 52 MMNCYLIKELLNTMLLWFIEIAFFMAGDS-PMIVIRRS------LSPPAMKLIRLLLLDF 104
++ +L +E++NTMLLW E FF G ++++ RS ++PP RL+ LD
Sbjct: 212 VLTTFLTREVVNTMLLWPTETVFFGVGVLLTVVMLIRSFLRFLQITPPT----RLVCLDI 379
Query: 105 CILPMFRLGHHLYGFWWHWIPAVTCFGYLVIVPVACAV*RHKMER*LILISMNLISHLQG 164
CIL MF LGH L+ H I VT G L IVP V ++ +R L+LISM +H
Sbjct: 380 CILQMFLLGHQLHHILLH*ILGVTYSGSLAIVPNVSMVYSYQQDRKLLLISMITKNHPLA 559
Query: 165 KMFHAIATCVNK-HNAIHP-VAVVGMKLSIFQMILHLLDS*LKMCYI*LQIM-IKQKILT 221
KM+ AIA V++ H+A+H VA+V +KL+I+Q IL LL S +MC *LQIM IK +LT
Sbjct: 560 KMWPAIAAYVSRRHSALHHLVALVHIKLNIYQKILQLLVSW*RMCCT*LQIMMIKLNMLT 739
Query: 222 HKLPLVVAKFRLVCF*MVQLQMVYLDLVWKMYLFRAS*PKRGLFQILFQCVLDLMVLEES 281
H L LVV KF+ F M QLQMV LD MYLF+ S*P +G QILFQCVL LM LEES
Sbjct: 740 H*LLLVVDKFKQGLFWMGQLQMVCLD*E*VMYLFQVS*PNKG*LQILFQCVLLLMALEES 919
Query: 282 RLG-IPAAQTKEKHHSTSGNHILHITSPLHKLLWEDMLPIMSFMQFSTPVPHSHT*MTQL 340
LG I A TKEKHHST G I TS KLLWE++ I S MQ+ST PH HT* T+L
Sbjct: 920 HLGIIIVAWTKEKHHSTLGRRIQLTTSLSPKLLWEEIPLIWSSMQYSTLAPHLHT*TTRL 1099
Query: 341 IR*FPRSSIL*SKQIDIRLCH--LIRISPLNTVMT*VQIRQLKFPF*I*Q*KVEMIIMSR 398
I + I +++ + H ++ I PL+T MT* Q R+LKFP *+*KVE II+S
Sbjct: 1100ISKLLKVLI---QKLSYKGIHFLILMIFPLSTAMT*EQTRRLKFPILT*R*KVETIILSW 1270
Query: 399 IQLYLFLVRLKEIFFVWEYRKVTI*ISLDKTS*LVTV*SSIVRT*TLVGRNLTVLKKCS- 457
+ + + +FFVW + K T+*ISLDKTS*LVT *S I RT* VGRNLTV+ S
Sbjct: 1271TR**Q-VAGVTMVFFVWLF*KATM*ISLDKTS*LVTG*SLIART*LWVGRNLTVMMMNSR 1447
Query: 458 PYLLTNLILLLFPLLLLLILWLDQTHQVILEDFHLISHLGRSLLLHL 504
YLLT+ LL IL + T Q++L+ FHL++HL ++L HL
Sbjct: 1448VYLLTDRTPQPCLLLWR*ILKYNLTLQMVLKGFHLVTHLRKNLH*HL 1588
>BF641229 weakly similar to GP|22655368|gb| At2g17760/At2g17760 {Arabidopsis
thaliana}, partial (16%)
Length = 465
Score = 287 bits (734), Expect = 8e-78
Identities = 130/131 (99%), Positives = 130/131 (99%)
Frame = +1
Query: 1 MTLYIRHWCCCCC*WCWC*VYLHTVATAWGNSVWIFIIDSRIQ*PRF*ALVMMNCYLIKE 60
MTLYIRHWCCCCC*WCWC*VYLHTVATAWGNSVWIFIIDSRIQ*PRF*ALVMMNCYLIKE
Sbjct: 73 MTLYIRHWCCCCC*WCWC*VYLHTVATAWGNSVWIFIIDSRIQ*PRF*ALVMMNCYLIKE 252
Query: 61 LLNTMLLWFIEIAFFMAGDSPMIVIRRSLSPPAMKLIRLLLLDFCILPMFRLGHHLYGFW 120
LLNTMLLWFIEIAFFMAGDSPMIVIRRSLSPPAMKLI LLLLDFCILPMFRLGHHLYGFW
Sbjct: 253 LLNTMLLWFIEIAFFMAGDSPMIVIRRSLSPPAMKLIXLLLLDFCILPMFRLGHHLYGFW 432
Query: 121 WHWIPAVTCFG 131
WHWIPAVTCFG
Sbjct: 433 WHWIPAVTCFG 465
>TC90871 similar to PIR|T45765|T45765 hypothetical protein F24M12.380 -
Arabidopsis thaliana, partial (7%)
Length = 507
Score = 276 bits (706), Expect = 1e-74
Identities = 149/149 (100%), Positives = 149/149 (100%)
Frame = +3
Query: 362 LIRISPLNTVMT*VQIRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVT 421
LIRISPLNTVMT*VQIRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVT
Sbjct: 24 LIRISPLNTVMT*VQIRQLKFPF*I*Q*KVEMIIMSRIQLYLFLVRLKEIFFVWEYRKVT 203
Query: 422 I*ISLDKTS*LVTV*SSIVRT*TLVGRNLTVLKKCSPYLLTNLILLLFPLLLLLILWLDQ 481
I*ISLDKTS*LVTV*SSIVRT*TLVGRNLTVLKKCSPYLLTNLILLLFPLLLLLILWLDQ
Sbjct: 204 I*ISLDKTS*LVTV*SSIVRT*TLVGRNLTVLKKCSPYLLTNLILLLFPLLLLLILWLDQ 383
Query: 482 THQVILEDFHLISHLGRSLLLHLW*SFLH 510
THQVILEDFHLISHLGRSLLLHLW*SFLH
Sbjct: 384 THQVILEDFHLISHLGRSLLLHLW*SFLH 470
>BF005656
Length = 149
Score = 92.8 bits (229), Expect = 3e-19
Identities = 43/49 (87%), Positives = 45/49 (91%)
Frame = +1
Query: 270 QCVLDLMVLEESRLGIPAAQTKEKHHSTSGNHILHITSPLHKLLWEDML 318
QCV DLMVLEESRLGIPAA T EKHHS+SGNH+LHIT PLHKLLWEDML
Sbjct: 1 QCV*DLMVLEESRLGIPAAHTNEKHHSSSGNHMLHITPPLHKLLWEDML 147
>TC84053 similar to PIR|T04698|T04698 hypothetical protein F4B14.150 -
Arabidopsis thaliana, partial (19%)
Length = 590
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/48 (50%), Positives = 26/48 (54%)
Frame = +1
Query: 88 SLSPPAMKLIRLLLLDFCILPMFRLGHHLYGFWWHWIPAVTCFGYLVI 135
SLSP A+ LDFCI+ GH WWH I VTCFGY VI
Sbjct: 286 SLSPTAIPPSVSAPLDFCIIQQLS*GHLE*SLWWHLIQEVTCFGYPVI 429
>BF646914 similar to GP|22655368|gb| At2g17760/At2g17760 {Arabidopsis
thaliana}, partial (7%)
Length = 651
Score = 37.4 bits (85), Expect = 0.014
Identities = 21/51 (41%), Positives = 27/51 (52%)
Frame = +1
Query: 99 LLLLDFCILPMFRLGHHLYGFWWHWIPAVTCFGYLVIVPVACAV*RHKMER 149
L+ + CIL MF LGH L+ H I VT G L IVP V ++ +R
Sbjct: 379 LVCISVCILQMFLLGHQLHHILLH*ILGVTYSGSLAIVPNVSMVYSYQQDR 531
>TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-binding protein
[imported] - Arabidopsis thaliana, partial (57%)
Length = 1695
Score = 35.4 bits (80), Expect = 0.053
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = -3
Query: 8 WCCCCC*WCWC* 19
WCCCCC CWC*
Sbjct: 517 WCCCCCCCCWC* 482
>TC87093 homologue to GP|10177817|dbj|BAB11183. gene_id:MKD15.14~unknown
protein {Arabidopsis thaliana}, partial (49%)
Length = 1313
Score = 35.0 bits (79), Expect = 0.069
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = -2
Query: 8 WCCCCC*WCWC 18
WCCCCC WC C
Sbjct: 670 WCCCCCCWCCC 638
>BQ751599 similar to GP|7293866|gb| ftz-f1 gene product {Drosophila
melanogaster}, partial (3%)
Length = 410
Score = 32.7 bits (73), Expect = 0.34
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = +1
Query: 8 WCCCCC*WCWC*VYLHTVAT 27
WCCC WCWC +H +++
Sbjct: 280 WCCCW*CWCWCNRVVHEISS 339
Score = 31.6 bits (70), Expect = 0.76
Identities = 9/12 (75%), Positives = 9/12 (75%), Gaps = 1/12 (8%)
Frame = +1
Query: 8 WCCCCC*W-CWC 18
W CCCC W CWC
Sbjct: 250 WLCCCCCW*CWC 285
>AJ388998 similar to SP|Q9SS17|RS24_ 40S ribosomal protein S24. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (70%)
Length = 538
Score = 32.3 bits (72), Expect = 0.45
Identities = 13/60 (21%), Positives = 31/60 (51%)
Frame = -2
Query: 260 PKRGLFQILFQCVLDLMVLEESRLGIPAAQTKEKHHSTSGNHILHITSPLHKLLWEDMLP 319
P + +F+++F C+L+ ++ + G+ ++ K + +LH +P LL ++P
Sbjct: 183 PNKSIFRLVFLCILNRIIDQTKTSGLATSKMSSKLEHKNSVWVLHXVNPCKLLLEFSLVP 4
>TC93889
Length = 433
Score = 32.3 bits (72), Expect = 0.45
Identities = 30/134 (22%), Positives = 61/134 (45%), Gaps = 11/134 (8%)
Frame = +3
Query: 78 GDSPMIVIRRSLSPPAMKLIRLLLLDFCILPMFRLGHHLYGFW-WHWIPA---------V 127
G S +I++ P + K+ R L +C+LP+ +L +L+GF+ W+ + +
Sbjct: 27 GMSFLIMMNCMECPNSHKVFRRLYFLYCLLPVLQLLAYLFGFFIWNRVTVLGKHTSSWLI 206
Query: 128 TCFGYLVIVPVACAV*RHKMER*LILISM-NLISHLQGKMFHAIATCVNKHNAIHPVAVV 186
TC+ + V A+ H +L+S+ N+I+ + H C N P + +
Sbjct: 207 TCYLNTYVSDVIHAI--HICSLPSVLVSI*NVIADSDLYVVHLFGLCCNYLEYYKPWSSI 380
Query: 187 GMKLSIFQMILHLL 200
+ + + LH++
Sbjct: 381 CLDGFKYNLQLHVI 422
>TC87569 similar to PIR|E96776|E96776 hypothetical protein F25A4.30
[imported] - Arabidopsis thaliana, partial (12%)
Length = 885
Score = 32.3 bits (72), Expect = 0.45
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = +2
Query: 9 CCCCC*WCW 17
CCCCC WCW
Sbjct: 656 CCCCCSWCW 682
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 32.3 bits (72), Expect = 0.45
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = -1
Query: 9 CCCCC*WCW 17
CCCCC WCW
Sbjct: 285 CCCCCCWCW 259
>AW685128 homologue to GP|1556394|emb| GATA factor {Drosophila melanogaster},
partial (2%)
Length = 634
Score = 31.6 bits (70), Expect = 0.76
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = -3
Query: 9 CCCCC*WCWC 18
CCCCC W WC
Sbjct: 194 CCCCCCWWWC 165
>TC85402 similar to GP|21554238|gb|AAM63313.1 Contains similarity to bHLH
transcription factor GBOF-1 from Tulipa gesneriana
gb|AF185269, partial (32%)
Length = 1410
Score = 31.6 bits (70), Expect = 0.76
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = -2
Query: 9 CCCCC*WCWC 18
CCCCC W WC
Sbjct: 290 CCCCCCWWWC 261
>TC86767 similar to PIR|T52470|T52470 cytochrome-b5 reductase (EC 1.6.2.2)
[validated] - Arabidopsis thaliana, partial (88%)
Length = 1174
Score = 31.2 bits (69), Expect = 1.00
Identities = 9/13 (69%), Positives = 9/13 (69%), Gaps = 2/13 (15%)
Frame = +3
Query: 8 WC--CCCC*WCWC 18
WC CCC WCWC
Sbjct: 147 WCRRCCCSHWCWC 185
>TC81797 similar to GP|17065224|gb|AAL32766.1 Unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 758
Score = 31.2 bits (69), Expect = 1.00
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = -3
Query: 8 WCCCCC*WCWC 18
+CCCCC WC C
Sbjct: 468 FCCCCCCWCCC 436
>TC83050 similar to SP|Q9FGZ4|WR48_ARATH Probable WRKY transcription
factor 48 (WRKY DNA-binding protein 48). [Mouse-ear
cress], partial (31%)
Length = 974
Score = 26.9 bits (58), Expect(2) = 1.2
Identities = 9/20 (45%), Positives = 10/20 (50%)
Frame = -1
Query: 10 CCCC*WCWC*VYLHTVATAW 29
CCCC*W W + H W
Sbjct: 83 CCCC*WWWWLLESHRRMPWW 24
Score = 22.3 bits (46), Expect(2) = 1.2
Identities = 6/6 (100%), Positives = 6/6 (100%)
Frame = -1
Query: 9 CCCCC* 14
CCCCC*
Sbjct: 221 CCCCC* 204
>BE997694 similar to GP|13646986|db DNA-binding protein DF1 {Pisum sativum},
partial (19%)
Length = 609
Score = 30.8 bits (68), Expect = 1.3
Identities = 9/13 (69%), Positives = 9/13 (69%), Gaps = 2/13 (15%)
Frame = -2
Query: 8 WCCCCC--*WCWC 18
WCCCC WCWC
Sbjct: 404 WCCCCWQRYWCWC 366
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.357 0.158 0.571
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,209,493
Number of Sequences: 36976
Number of extensions: 347565
Number of successful extensions: 10971
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 5253
Number of HSP's successfully gapped in prelim test: 335
Number of HSP's that attempted gapping in prelim test: 3198
Number of HSP's gapped (non-prelim): 7432
length of query: 513
length of database: 9,014,727
effective HSP length: 100
effective length of query: 413
effective length of database: 5,317,127
effective search space: 2195973451
effective search space used: 2195973451
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136504.3


