

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.11 + phase: 0
(137 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
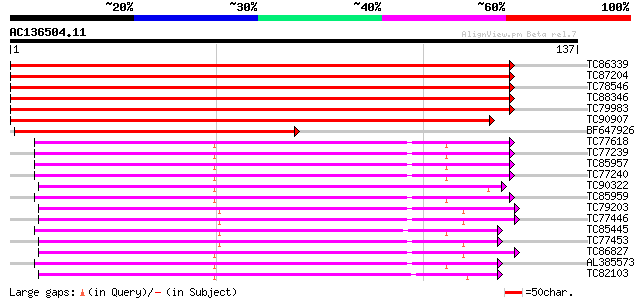
Score E
Sequences producing significant alignments: (bits) Value
TC86339 small G-protein ROP6 [Medicago truncatula] 245 3e-66
TC87204 small G-protein ROP9 [Medicago truncatula] 243 2e-65
TC78546 small G-protein ROP3 [Medicago truncatula] 238 6e-64
TC88346 homologue to SP|Q40220|RAC2_LOTJA RAC-like GTP binding p... 232 3e-62
TC79983 homologue to SP|Q41253|RACD_GOSHI RAC-like GTP binding p... 230 1e-61
TC90907 homologue to PIR|T08950|T08950 GTP binding protein Arac7... 211 6e-56
BF647926 homologue to PIR|T06679|T066 GTP-binding protein Arac8 ... 121 1e-28
TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13... 82 9e-17
TC77239 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japo... 82 9e-17
TC85957 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 ... 82 9e-17
TC77240 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japo... 82 9e-17
TC90322 similar to GP|1370172|emb|CAA98163.1 RAB1X {Lotus japoni... 80 2e-16
TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 ... 80 2e-16
TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-... 80 3e-16
TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding prote... 80 3e-16
TC85445 homologue to GP|1370180|emb|CAA98167.1 RAB5B {Lotus japo... 80 3e-16
TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding prote... 80 3e-16
TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding prote... 79 7e-16
AL385573 homologue to GP|11558647|em secretion related GTPase (... 78 1e-15
TC82103 similar to GP|1370144|emb|CAA98178.1 RAB11B {Lotus japon... 77 2e-15
>TC86339 small G-protein ROP6 [Medicago truncatula]
Length = 1195
Score = 245 bits (626), Expect = 3e-66
Identities = 120/122 (98%), Positives = 121/122 (98%)
Frame = +2
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD
Sbjct: 230 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 409
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL
Sbjct: 410 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 589
Query: 121 GM 122
+
Sbjct: 590 DL 595
>TC87204 small G-protein ROP9 [Medicago truncatula]
Length = 995
Score = 243 bits (620), Expect = 2e-65
Identities = 119/122 (97%), Positives = 120/122 (97%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGS VNLGLWD
Sbjct: 153 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSIVNLGLWD 332
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL
Sbjct: 333 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 512
Query: 121 GM 122
+
Sbjct: 513 DL 518
>TC78546 small G-protein ROP3 [Medicago truncatula]
Length = 1177
Score = 238 bits (607), Expect = 6e-64
Identities = 113/122 (92%), Positives = 120/122 (97%)
Frame = +1
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MS SRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV+GST+NLGLWD
Sbjct: 139 MSGSRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVDGSTINLGLWD 318
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVF+LAFSLISKASYEN++KKWIPEL+HYAPGVPIILVGTKL
Sbjct: 319 TAGQEDYNRLRPLSYRGADVFLLAFSLISKASYENIAKKWIPELRHYAPGVPIILVGTKL 498
Query: 121 GM 122
+
Sbjct: 499 DL 504
Score = 25.0 bits (53), Expect = 9.7
Identities = 13/29 (44%), Positives = 16/29 (54%)
Frame = -3
Query: 11 TVGDGAVGKTCLLISYTSNTFPTDYVPTV 39
TVG +VGK L+ + FPT PTV
Sbjct: 254 TVGT*SVGKVLLV*EISKQVFPTAPSPTV 168
>TC88346 homologue to SP|Q40220|RAC2_LOTJA RAC-like GTP binding protein
RAC2. {Lotus japonicus}, partial (77%)
Length = 703
Score = 232 bits (592), Expect = 3e-62
Identities = 109/122 (89%), Positives = 119/122 (97%)
Frame = +2
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MS +RFIKCVTVGDGAVGKTC+LISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWD
Sbjct: 248 MSTARFIKCVTVGDGAVGKTCMLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWD 427
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVF+LAFSL+S+ASYEN+SKKWIPEL+HYAP VPI+LVGTKL
Sbjct: 428 TAGQEDYNRLRPLSYRGADVFLLAFSLLSRASYENISKKWIPELRHYAPTVPIVLVGTKL 607
Query: 121 GM 122
+
Sbjct: 608 DL 613
>TC79983 homologue to SP|Q41253|RACD_GOSHI RAC-like GTP binding protein
RAC13. [Upland cotton] {Gossypium hirsutum}, complete
Length = 902
Score = 230 bits (587), Expect = 1e-61
Identities = 109/122 (89%), Positives = 117/122 (95%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MS +RFIKCVTVGDGAVGKTC+LISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWD
Sbjct: 132 MSTARFIKCVTVGDGAVGKTCMLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWD 311
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVF+L FSLISKASYEN+SKKWI EL+HYAP VPI+LVGTKL
Sbjct: 312 TAGQEDYNRLRPLSYRGADVFLLCFSLISKASYENISKKWISELRHYAPNVPIVLVGTKL 491
Query: 121 GM 122
+
Sbjct: 492 DL 497
>TC90907 homologue to PIR|T08950|T08950 GTP binding protein Arac7 [imported]
- Arabidopsis thaliana, partial (55%)
Length = 474
Score = 211 bits (538), Expect = 6e-56
Identities = 98/117 (83%), Positives = 110/117 (93%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSAS+FIKCVTVGDGAVGKTC+LI YTSN FPTDY+PTVFDNFSANV V+GS VNLGLWD
Sbjct: 123 MSASKFIKCVTVGDGAVGKTCMLICYTSNKFPTDYIPTVFDNFSANVAVDGSIVNLGLWD 302
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVG 117
TAGQEDY+RLRPLSYRGAD+F+LAFSLIS+ASYENV KKW+PEL+ +AP VPI+LVG
Sbjct: 303 TAGQEDYSRLRPLSYRGADIFVLAFSLISRASYENVLKKWMPELRRFAPNVPIVLVG 473
>BF647926 homologue to PIR|T06679|T066 GTP-binding protein Arac8 -
Arabidopsis thaliana, partial (31%)
Length = 646
Score = 121 bits (303), Expect = 1e-28
Identities = 57/69 (82%), Positives = 61/69 (87%)
Frame = +1
Query: 2 SASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWDT 61
+ASRFIKCVTVGDGAVGKTC+LI YTSN FPTDY+PTVFDNFSANVVV G TVNLGLWDT
Sbjct: 88 TASRFIKCVTVGDGAVGKTCMLICYTSNKFPTDYIPTVFDNFSANVVVEGITVNLGLWDT 267
Query: 62 AGQEDYNRL 70
AG Y+ L
Sbjct: 268 AGMFTYSLL 294
>TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13 - garden
pea, complete
Length = 1136
Score = 81.6 bits (200), Expect = 9e-17
Identities = 41/118 (34%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +2
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 263 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 442
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + +AS+ N+ + WI + +H + V ILVG K M
Sbjct: 443 RFRTITTAYYRGAMGILLVYDVTDEASFNNI-RNWIRNIEQHASDNVNKILVGNKADM 613
>TC77239 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japonicus},
partial (96%)
Length = 816
Score = 81.6 bits (200), Expect = 9e-17
Identities = 41/118 (34%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +1
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 241 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 420
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + +AS+ N+ + WI + +H + V ILVG K M
Sbjct: 421 RFRTITTAYYRGAMGILLVYDVTDEASFNNI-RNWIRNIEQHASDNVNKILVGNKADM 591
>TC85957 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 - garden
pea, complete
Length = 1148
Score = 81.6 bits (200), Expect = 9e-17
Identities = 41/118 (34%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +3
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 252 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 431
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + ++S+ N+ K WI + +H + V ILVG K M
Sbjct: 432 RFRTITTAYYRGAMGILLVYDVTDESSFNNI-KNWIRNIEQHASDNVNKILVGNKADM 602
>TC77240 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japonicus},
complete
Length = 1148
Score = 81.6 bits (200), Expect = 9e-17
Identities = 41/118 (34%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +1
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 268 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 447
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + +AS+ N+ + WI + +H + V ILVG K M
Sbjct: 448 RFRTITTAYYRGAMGILLVYDVTDEASFNNI-RNWIRNIEQHASDNVNKILVGNKADM 618
>TC90322 similar to GP|1370172|emb|CAA98163.1 RAB1X {Lotus japonicus},
partial (98%)
Length = 1054
Score = 80.5 bits (197), Expect = 2e-16
Identities = 42/116 (36%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Frame = +1
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+++ SN+ D PT+ +F ++ V G + L +WDTAGQE
Sbjct: 295 KILLIGDSGVGKSCLLVTFISNSSVEDLSPTIGVDFKIKMLTVGGKRLKLTIWDTAGQER 474
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPII--LVGTKL 120
+ L YRGA IL + + + ++ N+S+ W EL+ Y+ + LVG K+
Sbjct: 475 FRTLTSSYYRGAQGIILVYDVTRRETFTNLSEVWSKELELYSTNQECVKMLVGNKV 642
>TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 - garden
pea, complete
Length = 886
Score = 80.5 bits (197), Expect = 2e-16
Identities = 40/118 (33%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +3
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 117 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 296
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + ++S+ N+ + WI + +H + V ILVG K M
Sbjct: 297 RFRTITTAYYRGAMGILLVYDVTDESSFNNI-RNWIRNIEQHASDNVNKILVGNKADM 467
>TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-like (clone
vfa-ypt1) - fava bean, complete
Length = 920
Score = 80.1 bits (196), Expect = 3e-16
Identities = 41/118 (34%), Positives = 68/118 (56%), Gaps = 2/118 (1%)
Frame = +1
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V +G T+ L +WDTAGQE
Sbjct: 235 KLLLIGDSGVGKSCLLLRFADDSYIDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQER 414
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K ++
Sbjct: 415 FRTITSSYYRGAHGIIIVYDVTDEESFNNV-KQWLSEIDRYASDNVNKLLVGNKCDLT 585
>TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding protein {Pisum
sativum}, complete
Length = 1080
Score = 80.1 bits (196), Expect = 3e-16
Identities = 42/118 (35%), Positives = 68/118 (57%), Gaps = 2/118 (1%)
Frame = +3
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V +G T+ L +WDTAGQE
Sbjct: 144 KLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQER 323
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K +S
Sbjct: 324 FRTITSSYYRGAHGIIVVYDVTDQESFNNV-KQWLNEIDRYASENVNKLLVGNKSDLS 494
>TC85445 homologue to GP|1370180|emb|CAA98167.1 RAB5B {Lotus japonicus},
partial (98%)
Length = 1147
Score = 80.1 bits (196), Expect = 3e-16
Identities = 43/116 (37%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Frame = +1
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV--NGSTVNLGLWDTAGQ 64
+K V +GD VGK+C+++ + F TV +F + + + +TV +WDTAGQ
Sbjct: 283 VKLVLLGDSGVGKSCIVLRFVRGQFDPTSKVTVGASFLSQTIALQDSTTVKFEIWDTAGQ 462
Query: 65 EDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTK 119
E Y L PL YRGA V ++A+ + S S+ ++ W+ EL KH +P + + LVG K
Sbjct: 463 ERYAALAPLYYRGAAVAVIAYDITSPESFSK-AQYWVKELQKHGSPDIVMALVGNK 627
>TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding protein {Pisum
sativum}, complete
Length = 944
Score = 79.7 bits (195), Expect = 3e-16
Identities = 41/114 (35%), Positives = 66/114 (56%), Gaps = 2/114 (1%)
Frame = +1
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V +G T+ L +WDTAGQE
Sbjct: 151 KLLLIGDSGVGKSCLLLRFADDSYIDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQER 330
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTK 119
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K
Sbjct: 331 FRTITSSYYRGAHGIIIVYDVTDEESFNNV-KQWLSEIDRYASDNVNKLLVGNK 489
>TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding protein {Pisum
sativum}, complete
Length = 917
Score = 78.6 bits (192), Expect = 7e-16
Identities = 41/118 (34%), Positives = 67/118 (56%), Gaps = 2/118 (1%)
Frame = +2
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQED 66
K + +GD +VGK+CLL+ + +++ Y+ T+ +F V + G T L +WDTAGQE
Sbjct: 152 KLLIIGDSSVGKSCLLLRFADDSYDDTYISTIGVDFKIRTVELEGKTAKLQIWDTAGQER 331
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + S+ NV K+W+ E+ YA V +LVG K ++
Sbjct: 332 FRTITSSYYRGAHGIIIVYDVTDIESFNNV-KQWLHEIDRYANHSVSKLLVGNKCDLT 502
>AL385573 homologue to GP|11558647|em secretion related GTPase (SrgA)
{Aspergillus niger}, partial (61%)
Length = 463
Score = 77.8 bits (190), Expect = 1e-15
Identities = 37/115 (32%), Positives = 68/115 (58%), Gaps = 2/115 (1%)
Frame = +2
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ ++F ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 92 IKLLLIGDSGVGKSCLLLRFSDDSFTPSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 271
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTK 119
+ + YRGA +L + + + S++N+ + W + +H + GV IL+G K
Sbjct: 272 RFRTITTAYYRGAMGILLVYDVTDERSFKNI-RNWFSNIEQHASEGVNKILIGNK 433
>TC82103 similar to GP|1370144|emb|CAA98178.1 RAB11B {Lotus japonicus},
partial (80%)
Length = 1513
Score = 77.0 bits (188), Expect = 2e-15
Identities = 43/114 (37%), Positives = 65/114 (56%), Gaps = 2/114 (1%)
Frame = +1
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQED 66
K V +GD AVGKT +L ++ N F D T+ F V +NG + +WDTAGQE
Sbjct: 220 KVVVIGDSAVGKTQILSRFSKNEFCFDSKSTIGVEFQTRTVTINGKVIKAQIWDTAGQER 399
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAP-GVPIILVGTK 119
Y + YRGA +L + + + S+++V+ KW+ EL+ +A + I+LVG K
Sbjct: 400 YRAVTSAYYRGALGAMLVYDITKRQSFDHVA-KWVEELRAHADNSIVIMLVGNK 558
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,826,749
Number of Sequences: 36976
Number of extensions: 62417
Number of successful extensions: 423
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 378
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 378
length of query: 137
length of database: 9,014,727
effective HSP length: 86
effective length of query: 51
effective length of database: 5,834,791
effective search space: 297574341
effective search space used: 297574341
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC136504.11


