

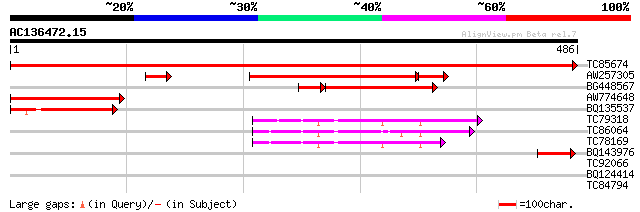
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
(486 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85674 similar to PIR|E96500|E96500 probable histidine decarbox... 997 0.0
AW257305 similar to GP|4996105|dbj serine decarboxylase {Brassic... 293 5e-91
BG448567 weakly similar to PIR|E96500|E96 probable histidine dec... 155 6e-44
AW774648 159 2e-39
BQ135537 weakly similar to PIR|E96500|E965 probable histidine de... 107 8e-24
TC79318 similar to PIR|H84431|H84431 probable glutamate decarbox... 56 3e-08
TC86064 similar to SP|P54767|DCE_LYCES Glutamate decarboxylase (... 52 4e-07
TC78169 homologue to SP|Q07346|DCE_PETHY Glutamate decarboxylase... 49 3e-06
BQ143976 45 8e-05
TC92066 similar to GP|12231296|gb|AAG49032.1 ripening regulated ... 31 1.2
BQ124414 similar to GP|6560753|gb|A F3M18.15 {Arabidopsis thalia... 28 6.0
TC84794 similar to PIR|S54156|S54156 extensin-like protein - cow... 28 7.9
>TC85674 similar to PIR|E96500|E96500 probable histidine decarboxylase
[imported] - Arabidopsis thaliana, partial (85%)
Length = 1839
Score = 997 bits (2578), Expect = 0.0
Identities = 486/486 (100%), Positives = 486/486 (100%)
Frame = +3
Query: 1 MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKRE 60
MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKRE
Sbjct: 108 MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKRE 287
Query: 61 IVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDY 120
IVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDY
Sbjct: 288 IVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDY 467
Query: 121 GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG 180
GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG
Sbjct: 468 GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG 647
Query: 181 NLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLR 240
NLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLR
Sbjct: 648 NLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLR 827
Query: 241 HQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAP 300
HQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAP
Sbjct: 828 HQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAP 1007
Query: 301 KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHA 360
KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHA
Sbjct: 1008KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHA 1187
Query: 361 PIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEF 420
PIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEF
Sbjct: 1188PIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEF 1367
Query: 421 IRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSC 480
IRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSC
Sbjct: 1368IRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSC 1547
Query: 481 LCAQHK 486
LCAQHK
Sbjct: 1548LCAQHK 1565
>AW257305 similar to GP|4996105|dbj serine decarboxylase {Brassica napus},
partial (39%)
Length = 584
Score = 293 bits (751), Expect(2) = 5e-91
Identities = 145/147 (98%), Positives = 146/147 (98%)
Frame = +3
Query: 206 SIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDL 265
SIFKAARMYRMECEKVETL+SGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDL
Sbjct: 66 SIFKAARMYRMECEKVETLSSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDL 245
Query: 266 VIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPC 325
VIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPC
Sbjct: 246 VIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPC 425
Query: 326 GVQITRLEHINALSRNVEYLASRDATI 352
GVQITRLEHIN LSRNVEYLASRDATI
Sbjct: 426 GVQITRLEHINGLSRNVEYLASRDATI 506
Score = 59.7 bits (143), Expect(2) = 5e-91
Identities = 25/26 (96%), Positives = 26/26 (99%)
Frame = +1
Query: 351 TIMGSRNGHAPIFLWYTLNRKGYRGF 376
+IMGSRNGHAPIFLWYTLNRKGYRGF
Sbjct: 505 SIMGSRNGHAPIFLWYTLNRKGYRGF 582
Score = 48.9 bits (115), Expect = 4e-06
Identities = 21/22 (95%), Positives = 21/22 (95%)
Frame = +2
Query: 117 DFDYGALSQLQHFSINNLGDPF 138
DFDYGALSQL HFSINNLGDPF
Sbjct: 2 DFDYGALSQLHHFSINNLGDPF 67
>BG448567 weakly similar to PIR|E96500|E96 probable histidine decarboxylase
[imported] - Arabidopsis thaliana, partial (24%)
Length = 361
Score = 155 bits (393), Expect(2) = 6e-44
Identities = 66/96 (68%), Positives = 83/96 (85%)
Frame = +2
Query: 271 EEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQIT 330
EE+GFS+DRFYIH DGALFG+M+P +K+AP +TFKKPIGS+++SGHKF+GCP+PCGV +T
Sbjct: 71 EESGFSRDRFYIHCDGALFGMMLPLLKQAPSITFKKPIGSITISGHKFLGCPIPCGVLLT 250
Query: 331 RLEHINALSRNVEYLASRDATIMGSRNGHAPIFLWY 366
RL HIN L ++VE + SRD TI G +GHAPIFLWY
Sbjct: 251 RLXHINTLCKDVEVIGSRDTTISGXXSGHAPIFLWY 358
Score = 40.0 bits (92), Expect(2) = 6e-44
Identities = 17/23 (73%), Positives = 22/23 (94%)
Frame = +1
Query: 248 INVNIGTTVKGAVDDLDLVIQKL 270
IN+NIGTTVKGA+D++D+VIQ L
Sbjct: 1 INLNIGTTVKGAIDNIDIVIQTL 69
>AW774648
Length = 359
Score = 159 bits (402), Expect = 2e-39
Identities = 80/98 (81%), Positives = 86/98 (87%)
Frame = +3
Query: 1 MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKRE 60
M SA V VNG STNGA+ELLPDD DVSAIIKDP PPVVAADNGIGKEEAK+NGGK+K+E
Sbjct: 66 MGASAHVFVNGSSTNGALELLPDDVDVSAIIKDPEPPVVAADNGIGKEEAKVNGGKKKKE 245
Query: 61 IVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARY 98
I +G +IHTTCL VTEPEADDEI GDRDAHMASVL RY
Sbjct: 246 IDIGIDIHTTCL*VTEPEADDEIAGDRDAHMASVLDRY 359
>BQ135537 weakly similar to PIR|E96500|E965 probable histidine decarboxylase
[imported] - Arabidopsis thaliana, partial (6%)
Length = 844
Score = 107 bits (268), Expect = 8e-24
Identities = 63/96 (65%), Positives = 69/96 (71%), Gaps = 4/96 (4%)
Frame = +3
Query: 1 MVGSADVLVNGLS----TNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGK 56
MVGSA VLVNGL T ++PD VS+IIKDP PPV AA N +GKEEA INGGK
Sbjct: 114 MVGSAHVLVNGLDH*WITPNYCLMIPD---VSSIIKDPEPPVPAAHNCMGKEEA*INGGK 284
Query: 57 EKREIVLGRNIHTTCLEVTEPEADDEITGDRDAHMA 92
EK +I LGRNIHTT LE T PEA DE T DR+AHMA
Sbjct: 285 EKIDIFLGRNIHTTWLEATAPEAYDECTRDRNAHMA 392
>TC79318 similar to PIR|H84431|H84431 probable glutamate decarboxylase
[imported] - Arabidopsis thaliana, partial (77%)
Length = 1934
Score = 56.2 bits (134), Expect = 3e-08
Identities = 53/206 (25%), Positives = 99/206 (47%), Gaps = 9/206 (4%)
Frame = +1
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E +V+ + D KA + ++ + + +G+T G +D+ D
Sbjct: 604 KFARYFEVELREVKVREDYYV-MDPAKAVEMVDENTICVAAI-LGSTYNGEFEDVKLLND 777
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKF--VGC 321
L+++K ++ G+ IHVD A G + PF+ + F+ P + S++VSGHK+ V
Sbjct: 778 LLLEKNKQTGWDTP---IHVDAASGGFIAPFLYPELEWDFRLPLVKSINVSGHKYGLVYA 948
Query: 322 PMPCGVQITRLEHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
+ + T+ + L ++ YL + T+ S+ I +Y L R G G++
Sbjct: 949 GIGWVIWRTKEDLPEELVFHINYLGADQPTFTLNFSKGSSQIIAQYYQLIRLGQEGYRSI 1128
Query: 380 VQKCLRNAHYFKDRLIEAGIGAMLNE 405
++ C NA K+ L E G +L++
Sbjct: 1129MEDCRENAMVLKEHLEETGYFNILSK 1206
>TC86064 similar to SP|P54767|DCE_LYCES Glutamate decarboxylase (EC
4.1.1.15) (GAD) (ERT D1). [Tomato] {Lycopersicon
esculentum}, partial (87%)
Length = 1759
Score = 52.4 bits (124), Expect = 4e-07
Identities = 53/201 (26%), Positives = 95/201 (46%), Gaps = 11/201 (5%)
Frame = +1
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L D KA + ++ + + +G+T+ G +D+ +
Sbjct: 586 KFARYFEVELKEVK-LTEDYYVMDPLKAVEMVDENTICVAAI-LGSTLTGEFEDVKLLNE 759
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+ +K +E G+ IHVD A G + PF+ F+ P + S++VSGHK+ G
Sbjct: 760 LLTKKNKETGWDTP---IHVDAASGGFVAPFLYPDLLWDFRLPLVKSINVSGHKY-GLVY 927
Query: 324 PCGVQITRLEH----INALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQ 377
P G+ H + L ++ YL S T+ S+ I +Y R G+ G++
Sbjct: 928 P-GIGWVVWRHKADLPDDLVFHINYLGSDQPTFTLNFSKGSSQIIAQYYQFIRLGFEGYK 1104
Query: 378 KEVQKCLRNAHYFKDRLIEAG 398
++ CL N K+ + + G
Sbjct: 1105NVMENCLANTRTLKEGIEKTG 1167
>TC78169 homologue to SP|Q07346|DCE_PETHY Glutamate decarboxylase (EC
4.1.1.15) (GAD). [Petunia] {Petunia hybrida}, partial
(67%)
Length = 1050
Score = 49.3 bits (116), Expect = 3e-06
Identities = 47/174 (27%), Positives = 87/174 (49%), Gaps = 9/174 (5%)
Frame = +3
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L+ G D KA L ++ + + +G+T+ G +D+ D
Sbjct: 543 KFARYFEVELKEVK-LSEGYYVMDPAKAVELVDENTICVAAI-LGSTLNGEFEDVKRLND 716
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKF--VGC 321
L+++K +E G+ IHVD A G + PF+ + F+ P + S++VSGHK+ V
Sbjct: 717 LLVEKNKETGWDTP---IHVDAASGGFIAPFIYPELEWDFRLPLVKSINVSGHKYGLVYA 887
Query: 322 PMPCGVQITRLEHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGY 373
+ + ++ + L ++ YL + T+ ++ I +Y L R GY
Sbjct: 888 GIGWAIWRSKEDLPEELIFHINYLGADQPTFTLNFXKSSSQVIAQYYQLIRLGY 1049
>BQ143976
Length = 446
Score = 44.7 bits (104), Expect = 8e-05
Identities = 16/33 (48%), Positives = 25/33 (75%)
Frame = +1
Query: 453 LVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
L++K++ W++D FQP CIA+DVG +C C+ H
Sbjct: 91 LLEKQSIWYKDEQFQPPCIANDVGSRNCACSIH 189
>TC92066 similar to GP|12231296|gb|AAG49032.1 ripening regulated protein
DDTFR18 {Lycopersicon esculentum}, partial (34%)
Length = 668
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +2
Query: 466 FQPYCIASDVGENSCLC 482
FQPYCI DVG +S LC
Sbjct: 251 FQPYCILYDVGHHSILC 301
>BQ124414 similar to GP|6560753|gb|A F3M18.15 {Arabidopsis thaliana}, partial
(6%)
Length = 626
Score = 28.5 bits (62), Expect = 6.0
Identities = 25/93 (26%), Positives = 43/93 (45%)
Frame = +3
Query: 89 AHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSR 148
AH+ SVL + K LTER Y + ++ + L H +++++ D + + + +
Sbjct: 120 AHLESVLEQTNKKLTERDLYLQEFENRINHISDKIHHL-HSTLSSIKDDSLHAETRIKAL 296
Query: 149 QFEVGVLDWFARLWELEKNEYWGYITNCGTEGN 181
EV +L W A L KN + +I E N
Sbjct: 297 DEEVQLL-WDA----LRKNNFDLHILKSKAEDN 380
>TC84794 similar to PIR|S54156|S54156 extensin-like protein - cowpea
(fragment), partial (49%)
Length = 478
Score = 28.1 bits (61), Expect = 7.9
Identities = 18/54 (33%), Positives = 23/54 (42%)
Frame = +1
Query: 62 VLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYN 115
+L + + T CL D I G HMA LA S T + GYPY+
Sbjct: 82 ILSQTLATNCLLPMTARGSDPIRGRFRPHMAMALAIVLISTTVVSVAADGYPYS 243
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,713,595
Number of Sequences: 36976
Number of extensions: 196175
Number of successful extensions: 826
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 822
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 825
length of query: 486
length of database: 9,014,727
effective HSP length: 100
effective length of query: 386
effective length of database: 5,317,127
effective search space: 2052411022
effective search space used: 2052411022
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136472.15


