

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136451.6 + phase: 0 /pseudo
(210 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
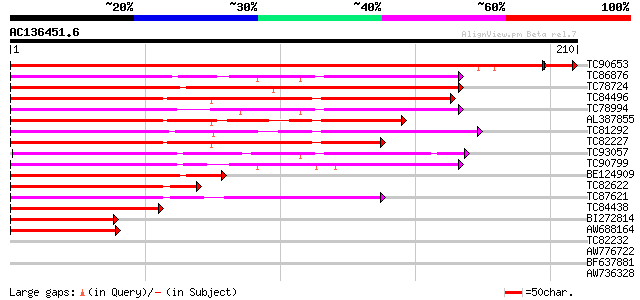
Score E
Sequences producing significant alignments: (bits) Value
TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populu... 377 e-107
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 112 8e-26
TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box t... 111 2e-25
TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 109 7e-25
TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS O... 103 6e-23
AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula... 102 8e-23
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 100 4e-22
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 98 3e-21
TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein ... 98 3e-21
TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcript... 95 2e-20
BE124909 similar to GP|4322475|gb| putative MADS box transcripti... 84 3e-17
TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {O... 82 2e-16
TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE pr... 81 3e-16
TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Be... 80 4e-16
BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdr... 66 8e-12
AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus... 60 8e-10
TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protei... 37 0.007
AW776722 weakly similar to GP|4008010|gb|A receptor-like protein... 30 0.51
BF637881 weakly similar to GP|20466530|gb| protein kinase-like p... 30 0.51
AW736328 similar to GP|7110148|gb| DNA repair-recombination prot... 30 0.51
>TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populus
balsamifera subsp. trichocarpa}, partial (59%)
Length = 845
Score = 377 bits (967), Expect(2) = e-107
Identities = 193/219 (88%), Positives = 195/219 (88%), Gaps = 20/219 (9%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP
Sbjct: 44 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 223
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL
Sbjct: 224 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 403
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL----VRTL 176
SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL +
Sbjct: 404 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELEKCVIHPQ 583
Query: 177 FL----------------SSYLYAFCQHHSHLNLPHHHG 199
FL +S LYAFCQHHSHLNLPHHHG
Sbjct: 584 FLFHDEGDEESAVALANGASTLYAFCQHHSHLNLPHHHG 700
Score = 27.3 bits (59), Expect(2) = e-107
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 199 GEEGYKNDDLRL 210
GEE YKNDDLRL
Sbjct: 699 GEESYKNDDLRL 734
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus
x domestica}, partial (85%)
Length = 2050
Score = 112 bits (281), Expect = 8e-26
Identities = 70/176 (39%), Positives = 105/176 (58%), Gaps = 8/176 (4%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+L++FS +++EY
Sbjct: 89 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEYANN 268
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLE-NLKKLKDINHKLRRQI-------RHRIGE 112
S + I++Y+K ++ E + E N + + + KLRRQI RH +GE
Sbjct: 269 --SVRATIERYKKACAA----STNAESVSEANTQFYQQESSKLRRQIRDIQNLNRHILGE 430
Query: 113 GGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
L LS ++L++LE + ++++R RK + + +K+ L+ N L
Sbjct: 431 A---LGSLSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNNYL 589
>TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box
transcription factor ETL {Eucalyptus globulus subsp.
globulus}, partial (71%)
Length = 1089
Score = 111 bits (277), Expect = 2e-25
Identities = 65/169 (38%), Positives = 103/169 (60%), Gaps = 1/169 (0%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK ++K IEN +NRQVT+SKRRNG+ KKA ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 226 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEFSSS 405
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLK-DINHKLRRQIRHRIGEGGMELDD 119
+S K +++YQ + ++ L ++ ++LK+ D KL + G EL
Sbjct: 406 SIS--KTVERYQGKVKELVLSTKGIQENTQHLKECDIDTTKKLEHLELSKRKLLGEELGS 579
Query: 120 LSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+F +L+ +E + S++KIR RK ++K + + + K R L + N L
Sbjct: 580 CAFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRL 726
>TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (74%)
Length = 681
Score = 109 bits (273), Expect = 7e-25
Identities = 63/169 (37%), Positives = 106/169 (62%), Gaps = 4/169 (2%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKAHE+SVLCDA+V+LI+FS K++EY +
Sbjct: 26 MGRGRVQLKKIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSNKGKLYEY-SS 202
Query: 61 GLSTKKIIDQYQK----TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
+KI+++Y++ + + E + +LK +++ R+ +GE E
Sbjct: 203 DPCMEKILERYERYSYAERQHVANDQPQNENWIIEHARLKTRLEVIQKNQRNFMGE---E 373
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMN 165
LD LS ++L+ LE ++S++ +IR RK ++ KK ++L++ N
Sbjct: 374 LDGLSMKELQHLEHQLDSALKQIRSRKNQLMYESISELSKKDKALQEKN 520
>TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS
OVEREXPRESSION 1 protein (Agamous-like MADS box protein
AGL20)., partial (83%)
Length = 946
Score = 103 bits (256), Expect = 6e-23
Identities = 66/183 (36%), Positives = 103/183 (56%), Gaps = 15/183 (8%)
Frame = +3
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK ++K IEN T+RQVT+SKRRNG+ KKA ELSVLCDA+V+LI+FS +++E+ +
Sbjct: 54 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSPRGRLYEFASS 233
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSH--------YEKMLENLKKLKDINHKLRRQI------ 106
+ L I+ +RSH + +EN ++LK+ + ++I
Sbjct: 234 SI------------LETIERYRSHTRINNTPTTSESVENTQQLKEEAENMMKKIDLLETS 377
Query: 107 -RHRIGEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMN 165
R +GEG L S +L+ +E+ + SI KIR +K V + + D ++K ++L N
Sbjct: 378 KRKLLGEG---LGSCSIDELQKIEQQLEKSINKIRVKKTKVFREQIDQLKEKEKALVAEN 548
Query: 166 GNL 168
L
Sbjct: 549 VRL 557
>AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula}, partial
(51%)
Length = 493
Score = 102 bits (255), Expect = 8e-23
Identities = 61/159 (38%), Positives = 100/159 (62%), Gaps = 12/159 (7%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN T++QVT+ KRR G+ KKA+E+SVLCDA+V+LIMFS K+ EY +
Sbjct: 53 MGRGRVQLKRIENKTSQQVTFFKRRTGLLKKANEISVLCDAQVALIMFSTKGKLFEY-SS 229
Query: 61 GLSTKKIIDQYQK------------TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRH 108
S + I+++Y++ T G+ W Y K+ ++ L+ R +R+
Sbjct: 230 APSMEDILERYERQNHTELTGATNETQGN---WSFEYMKLTAKVQVLE-------RNLRN 379
Query: 109 RIGEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVI 147
+G +LD LS ++L+SLE+ +++S+ +IR RK V+
Sbjct: 380 FVGN---DLDPLSVKELQSLEQQLDTSLKRIRTRKNQVM 487
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 100 bits (249), Expect = 4e-22
Identities = 61/185 (32%), Positives = 100/185 (53%), Gaps = 10/185 (5%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKI+I+ I+N T+RQVT+SKRRNG+ KKA EL++LCDA+V +++FS K++++ +
Sbjct: 71 MGRGKIQIRRIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVMIFSSTAKLYDFAS- 247
Query: 61 GLSTKKIIDQYQKT----------LGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRI 110
S + +ID+Y KT +I W+ + + L H L+ R +
Sbjct: 248 -TSMRSVIDRYNKTKEEHNQLGSSTSEIKFWQREAAMLRQQL-------HNLQESHRQIM 403
Query: 111 GEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 170
GE EL L+ ++L+ LE + S+ +R +K + +K + Q N L
Sbjct: 404 GE---ELSGLTVKELQGLENQLEISLRGVRMKKEQLFMDEIQELNRKGDIIHQENVELYR 574
Query: 171 ELVRT 175
++ T
Sbjct: 575 KVYGT 589
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 97.8 bits (242), Expect = 3e-21
Identities = 58/143 (40%), Positives = 93/143 (64%), Gaps = 4/143 (2%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKA E+SVLCDA+V+LI+FS K+ EY +
Sbjct: 130 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEY-SS 306
Query: 61 GLSTKKIIDQYQK----TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
+KI+++Y++ + +S E + KLK L+R R+ +GE +
Sbjct: 307 DPCMEKILERYERCSYMERQLVTSEQSPNENWVLEHAKLKARMEVLQRNQRNFMGE---D 477
Query: 117 LDDLSFQQLRSLEEDMNSSIAKI 139
LD L ++L+SLE+ ++S++ +I
Sbjct: 478 LDGLGLKELQSLEQQLDSALKQI 546
>TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein GHMADS-2
{Gossypium hirsutum}, partial (95%)
Length = 858
Score = 97.8 bits (242), Expect = 3e-21
Identities = 65/176 (36%), Positives = 99/176 (55%), Gaps = 7/176 (3%)
Frame = +3
Query: 2 GRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITPG 61
G ++ K IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY
Sbjct: 33 GGERLR*KGIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNN 212
Query: 62 LSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQI-------RHRIGEGG 114
+ + ID+Y+K D + E N + + + KLR+QI RH +G+
Sbjct: 213 I--RSTIDRYKKACSDHSSTTTTTE---INAQYYQQESAKLRQQIQMLQNSNRHLMGDA- 374
Query: 115 MELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 170
L L+ ++L+ LE + I +IR +K ++ + +K R +E N NL L
Sbjct: 375 --LSTLTVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQK--REIELENENLCL 530
>TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcription factor
{Canavalia lineata}, partial (76%)
Length = 653
Score = 94.7 bits (234), Expect = 2e-20
Identities = 64/175 (36%), Positives = 100/175 (56%), Gaps = 7/175 (4%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M R KI+IK I+N T RQVT+SKRR GIFKKA ELS+LCDA+V L++FS K++EY +
Sbjct: 71 MARQKIKIKKIDNATARQVTFSKRRRGIFKKAEELSILCDAEVGLVIFSTTGKLYEYASS 250
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLE---NLKKLKDINHKLRRQIRHRIGE-GGME 116
+ K II +Y + +SH+ L+ ++ K++ +L +++ R + GM+
Sbjct: 251 NM--KDIITRYGQ--------QSHHITKLDKPLQVQVEKNMPAELNKEVADRTQQLRGMK 400
Query: 117 LDD---LSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+D L+ + L+ LE+ + S + ++ E K I R K LE N +L
Sbjct: 401 SEDFEGLNLEGLQQLEKSLESXLKRVIEMKEKKILNEIKALRMKEIMLEXENKHL 565
>BE124909 similar to GP|4322475|gb| putative MADS box transcription factor
ETL {Eucalyptus globulus subsp. globulus}, partial (60%)
Length = 608
Score = 84.3 bits (207), Expect = 3e-17
Identities = 42/80 (52%), Positives = 61/80 (75%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK ++K IEN +NRQVT+SKRRNG+ KKA ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 148 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEFSSS 327
Query: 61 GLSTKKIIDQYQKTLGDIDL 80
+S K +++YQ + ++ L
Sbjct: 328 SIS--KTVERYQGKVKELVL 381
>TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {Oryza
sativa}, partial (78%)
Length = 847
Score = 81.6 bits (200), Expect = 2e-16
Identities = 39/71 (54%), Positives = 54/71 (75%)
Frame = +3
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKI I+ I+N T+RQVT+SKRR G+ KKA EL++LCDA+V L++FS K++EY
Sbjct: 138 MGRGKIVIRRIDNCTSRQVTFSKRRKGLIKKAKELAILCDAQVGLVIFSSTGKLYEY--A 311
Query: 61 GLSTKKIIDQY 71
S K +I++Y
Sbjct: 312 NTSMKSVIERY 344
>TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (65%)
Length = 1322
Score = 81.3 bits (199), Expect = 3e-16
Identities = 49/139 (35%), Positives = 79/139 (56%)
Frame = +2
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M R KI+IK IEN T RQVT+SKRR G+ KKA ELSVLCDA V+LI+FS K+ EY
Sbjct: 437 MAREKIQIKKIENSTARQVTFSKRRRGLIKKAEELSVLCDADVALIIFSSTGKLFEY--S 610
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
LS ++I++++ L + K+ E +L+ + + ++ + + +L +
Sbjct: 611 NLSMREILERHH-------LHSKNLAKLEEPSLELQLVENSNCSRLSKEVAQKSHQLRQM 769
Query: 121 SFQQLRSLEEDMNSSIAKI 139
+ L+ E ++I ++
Sbjct: 770 RGEDLQGFESGRVTTIGEV 826
>TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (24%)
Length = 702
Score = 80.5 bits (197), Expect = 4e-16
Identities = 36/57 (63%), Positives = 48/57 (84%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEY 57
MGRG++++K IEN NRQVT+SKRR+G+ KKA E+SVLCDA+V+LI+FS K+ EY
Sbjct: 223 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEY 393
>BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdragon,
partial (16%)
Length = 320
Score = 66.2 bits (160), Expect = 8e-12
Identities = 29/40 (72%), Positives = 36/40 (89%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCD 40
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCD
Sbjct: 199 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 318
>AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus grandis},
partial (31%)
Length = 549
Score = 59.7 bits (143), Expect = 8e-10
Identities = 27/41 (65%), Positives = 34/41 (82%)
Frame = +1
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDA 41
MGRG++ ++ IEN NRQVT+SKRR+G+ KKA EL VLCDA
Sbjct: 7 MGRGRVVLERIENKINRQVTFSKRRSGLLKKAFELCVLCDA 129
>TC82232 similar to GP|10177316|dbj|BAB10642. kinesin-like protein
{Arabidopsis thaliana}, partial (21%)
Length = 1181
Score = 36.6 bits (83), Expect = 0.007
Identities = 33/151 (21%), Positives = 67/151 (43%), Gaps = 16/151 (10%)
Frame = +3
Query: 25 RNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITPGLSTKKIIDQYQKTL--------- 75
+ G+F +E S CD K I + ++HE S ++ +D+ K L
Sbjct: 78 KTGLFGDGNEYSSDCDVKPVDITDVEPVEIHEKELEHSSAQQKLDRELKELDKKLEQKEA 257
Query: 76 -----GDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDLSFQQLRSLEE 130
+ + + HYEK L L+ K + +++ G+ +L + Q+L +LE
Sbjct: 258 EMKLVNNASVLKQHYEKKLNELEHEKKFLQREIEELKSTSGDSTHKLKEEYLQKLNALES 437
Query: 131 DMNSSIAKIRERKFHVI--KTRTDTCRKKVR 159
+ S + K ++ + H++ K + D K+++
Sbjct: 438 QV-SELKKKQDAQAHLLRQKQKGDEAAKRLQ 527
>AW776722 weakly similar to GP|4008010|gb|A receptor-like protein kinase
{Arabidopsis thaliana}, partial (6%)
Length = 590
Score = 30.4 bits (67), Expect = 0.51
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +3
Query: 19 VTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITPGLSTKKI 67
V ++KR+N KA EL + +S N+MHE + P L+ K+I
Sbjct: 93 VQFNKRKNNNLNKAKELGKYICSIMSTRTRMVTNQMHELLHPWLARKEI 239
>BF637881 weakly similar to GP|20466530|gb| protein kinase-like protein
{Arabidopsis thaliana}, partial (17%)
Length = 642
Score = 30.4 bits (67), Expect = 0.51
Identities = 15/24 (62%), Positives = 17/24 (70%)
Frame = -3
Query: 63 STKKIIDQYQKTLGDIDLWRSHYE 86
S K IID Y +LG I LWR+HYE
Sbjct: 313 SRKTIIDHYYCSLGKI-LWRTHYE 245
>AW736328 similar to GP|7110148|gb| DNA repair-recombination protein
{Arabidopsis thaliana}, partial (5%)
Length = 513
Score = 30.4 bits (67), Expect = 0.51
Identities = 21/66 (31%), Positives = 34/66 (50%)
Frame = +2
Query: 85 YEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKF 144
Y+ LENL+ LKD + LR I H E L D QQL +D+++ I + +
Sbjct: 125 YKLKLENLQTLKDAAYTLRENITHD-QEKAESLKD-QIQQLDGSIKDLDAKIDHVDKTLK 298
Query: 145 HVIKTR 150
H+++ +
Sbjct: 299 HLLELK 316
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,839,242
Number of Sequences: 36976
Number of extensions: 92749
Number of successful extensions: 936
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 892
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 930
length of query: 210
length of database: 9,014,727
effective HSP length: 92
effective length of query: 118
effective length of database: 5,612,935
effective search space: 662326330
effective search space used: 662326330
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC136451.6


