

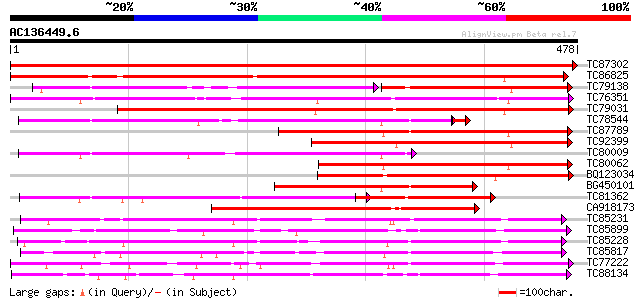
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136449.6 - phase: 0
(478 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 976 0.0
TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransfe... 410 e-115
TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 225 e-105
TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycos... 349 1e-96
TC79031 weakly similar to GP|13492674|gb|AAK28303.1 phenylpropan... 340 5e-94
TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 273 7e-75
TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 277 7e-75
TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose:... 257 6e-69
TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 244 7e-65
TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 243 9e-65
BQ123034 weakly similar to GP|14334982|gb putative glucosyltrans... 214 8e-56
BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosy... 205 4e-53
TC81362 similar to GP|19911201|dbj|BAB86927. glucosyltransferase... 202 2e-52
CA918173 weakly similar to GP|11034537|dbj putative glucosyl tra... 198 3e-51
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 197 1e-50
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 196 2e-50
TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 193 1e-49
TC85817 weakly similar to PIR|B85014|B85014 probable flavonol gl... 192 3e-49
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 191 4e-49
TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 191 5e-49
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 976 bits (2522), Expect = 0.0
Identities = 478/478 (100%), Positives = 478/478 (100%)
Frame = +1
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 60
MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN
Sbjct: 34 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 213
Query: 61 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM 120
IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM
Sbjct: 214 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM 393
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL 180
FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL
Sbjct: 394 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL 573
Query: 181 TKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLG 240
TKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLG
Sbjct: 574 TKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLG 753
Query: 241 PVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMG 300
PVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMG
Sbjct: 754 PVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMG 933
Query: 301 LEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGG 360
LEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGG
Sbjct: 934 LEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGG 1113
Query: 361 FVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPV 420
FVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPV
Sbjct: 1114FVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPV 1293
Query: 421 KKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSRAY 478
KKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSRAY
Sbjct: 1294KKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSRAY 1467
>TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransferase {Vigna
angularis}, partial (87%)
Length = 1704
Score = 410 bits (1053), Expect = e-115
Identities = 216/474 (45%), Positives = 300/474 (62%), Gaps = 3/474 (0%)
Frame = +3
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 60
M +E + I FFPF+ GH IP VD ARVF+ G TI+TT N +I + + +
Sbjct: 72 MNHETDAVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKS 251
Query: 61 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM 120
I + PE E +++ ++ I T +L EPL+ L Q +PD +V DM
Sbjct: 252 NHPITI---QILTTPENTEVTDTDMSAGPMI----DTSILLEPLKQFLVQHRPDGIVVDM 410
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL 180
F W+ D + IPRIVF+G G FP CV+ TR++ + +SS +EPF+VP LP I +
Sbjct: 411 FHRWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPGLPDIIEM 590
Query: 181 TKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLG 240
T+ Q P ++ F+ +++ E+ S G + N+FY+LEP YAD+ RN+LG+KAW +G
Sbjct: 591 TRSQTPIFMRNPSQFSDRIKQLEEN---SLGTLINSFYDLEPAYADYVRNKLGKKAWLVG 761
Query: 241 PVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMG 300
PVSLCNR E+K RG++ +IDE CL WL SK+PNSV+Y+ FGS+ QLKEIA G
Sbjct: 762 PVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYG 941
Query: 301 LEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGG 360
LEAS+ FIWVV K S +W+ + FE R++ KGLI RGWAPQ++IL+HE+VGG
Sbjct: 942 LEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGG 1121
Query: 361 FVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMG--GGE 418
F+THCGWNSTLEGV AG+PM TWP+ EQF N K ++D+++IGV VG + W E
Sbjct: 1122FMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKE 1301
Query: 419 PVKKDVIEKAVRRIMV-GDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIE 471
V ++ +E AV+++M ++ EEMR R K + A+RAVE GGSSY+D LI+
Sbjct: 1302LVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALIQ 1463
>TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (36%)
Length = 1575
Score = 225 bits (573), Expect(2) = e-105
Identities = 127/301 (42%), Positives = 174/301 (57%), Gaps = 9/301 (2%)
Frame = +2
Query: 20 HIIPCV-------DLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK-INIKTIKFPSPEE 71
H IPC D+A +F+S G VTI+TT N +I ++I + + T+ FPS +
Sbjct: 29 HSIPCTWPMTPLCDIATLFASCGHHVTIITTPSNAQIILKSIPSHNHLRLHTVPFPS-HQ 205
Query: 72 TGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWSTDSAAK 131
GLP G EN D K +T+LLR P+ H +EQ+ PDC+VAD F W + A +
Sbjct: 206 VGLPLGVENLAFVNNVDNSCKIHHATMLLRSPINHFVEQDSPDCIVADFMFLWVDELANR 385
Query: 132 FNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLPQLPQH 191
+IPR+ F+G F +C + K +D SS ++ LP ITL M
Sbjct: 386 LHIPRLAFNGFSLFAICAM---ESLKARDFESS-----IIQGLPHCITLNAMP------- 520
Query: 192 DKVFTKLLEESNESEVKSFGVIANTFYELE-PVYADHYRNELGRKAWHLGPVSLCNRDTE 250
K TK +E E+E+KS+G+I N F EL+ Y +HY +G +AWHLGP SL R T+
Sbjct: 521 PKALTKFMEPLLETELKSYGLIVNNFTELDGEEYIEHYEKTIGHRAWHLGPSSLICRTTQ 700
Query: 251 EKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIW 310
EKA RG+ + +D HECL WL SK+PNSV+Y+CFGS+ F++ QL EIA +EAS FIW
Sbjct: 701 EKADRGQTSVVDVHECLSWLNSKQPNSVLYICFGSLCHFTNKQLYEIASAIEASGHQFIW 880
Query: 311 V 311
V
Sbjct: 881 V 883
Score = 176 bits (445), Expect(2) = e-105
Identities = 88/165 (53%), Positives = 120/165 (72%), Gaps = 4/165 (2%)
Frame = +1
Query: 314 KSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEG 373
K +S EN +W+P+GFEER G+IIRGWAPQV+IL H ++G F+THCGWNST+E
Sbjct: 901 KEDESNDENEKWMPKGFEER----NIGMIIRGWAPQVVILGHPAIGAFLTHCGWNSTVEA 1068
Query: 374 VSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVK---KDVIEKAVR 430
VSAG+PM+TWP++ EQFYN K ++ + IGV VGV+ W +G + K +D+IEKA+R
Sbjct: 1069 VSAGVPMITWPVHDEQFYNEKLITQVRGIGVEVGVEEWSFIGFMKKKKIVGRDIIEKALR 1248
Query: 431 RIMVGD-EAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLK 474
R+M G EA E+R RA+E+ A+RAV+ GGSS+ + LI+DLK
Sbjct: 1249 RLMDGGIEAVEIRKRAQEYAIKAKRAVQEGGSSHKNLMALIDDLK 1383
>TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycose {Cicer
arietinum}, partial (92%)
Length = 1645
Score = 349 bits (896), Expect = 1e-96
Identities = 195/487 (40%), Positives = 292/487 (59%), Gaps = 12/487 (2%)
Frame = +2
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK-- 58
M E++ L I PF A GH+IP V+LAR+ +S+ VTI+TT N L +TI + K
Sbjct: 74 MGTESKPLKIYMLPFFAQGHLIPLVNLARLVASKNQHVTIITTPSNAQLFDKTIEEEKAA 253
Query: 59 ---INIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDC 115
I + IKFPS + GLP G EN +A K + ++ +E +++ PD
Sbjct: 254 GHHIRVHIIKFPSAQ-LGLPTGVENLFAASDNQTAGKIHMAAHFVKADIEEFMKENPPDV 430
Query: 116 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLP 175
++D+ F WS +A IPR+VF+ + F +C++ + + P+ VS + P+ + LP
Sbjct: 431 FISDIIFTWSESTAKNLQIPRLVFNPISIFDVCMIQAIQSH-PESFVSD-SGPYQIHGLP 604
Query: 176 GEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRK 235
+TL P F +L E E+E S GVI N+F EL+ Y ++Y N GRK
Sbjct: 605 HPLTLPIKPSPG-------FARLTESLIEAENDSHGVIVNSFAELDEGYTEYYENLTGRK 763
Query: 236 AWHLGPVSLCNRDTEEKACRGRE--ASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQ 293
WH+GP SL ++K E +SI +H+ L WL +KEP+SV+Y+ FGS+ S+ Q
Sbjct: 764 VWHVGPTSLMVEIPKKKKVVSTENDSSITKHQSLTWLDTKEPSSVLYISFGSLCRLSNEQ 943
Query: 294 LKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMIL 353
LKE+A G+EAS+ F+WVV + +N WLP+GF ER++ KG++I+GW PQ +IL
Sbjct: 944 LKEMANGIEASKHQFLWVVHGKEGEDEDN--WLPKGFVERMKEEKKGMLIKGWVPQALIL 1117
Query: 354 DHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIG 413
DH S+GGF+THCGWN+T+E +S+G+PMVT P +G+Q+YN K ++++ +IGV VG W
Sbjct: 1118DHPSIGGFLTHCGWNATVEAISSGVPMVTMPGFGDQYYNEKLVTEVHRIGVEVGAAEW-S 1294
Query: 414 MGGGEP----VKKDVIEKAVRRIMVGD-EAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
M + V+ + IEKAV+++M + E E+R RAKE + A +AV+ GGSS N +
Sbjct: 1295MSPYDAKKTVVRAERIEKAVKKLMDSNGEGGEIRKRAKEMKEKAWKAVQEGGSSQNCLTK 1474
Query: 469 LIEDLKS 475
L++ L S
Sbjct: 1475LVDYLHS 1495
>TC79031 weakly similar to GP|13492674|gb|AAK28303.1
phenylpropanoid:glucosyltransferase 1 {Nicotiana
tabacum}, partial (32%)
Length = 1385
Score = 340 bits (873), Expect = 5e-94
Identities = 177/391 (45%), Positives = 247/391 (62%), Gaps = 7/391 (1%)
Frame = +1
Query: 92 KFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLA 151
K +L+ +E + E +PD +V DMFFPWS D A K IPRI+FHG +
Sbjct: 28 KIYMGLYILQPDIEKLFETLQPDFIVTDMFFPWSADVAKKLGIPRIMFHGASYLARSAAH 207
Query: 152 CTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFG 211
Y+P K S T+ FV+P+LP E+ +T++QLP + + +L++ ESE KSFG
Sbjct: 208 SVEVYRPHLKAESDTDKFVIPDLPDELEMTRLQLPDWLRSPNQYAELMKVIKESEKKSFG 387
Query: 212 VIANTFYELEPVYADHYRNELGRKAWHLGPVSL-CNRDTEEKACRG---REASIDEHECL 267
+ N+FY+LE Y DHY+ +G K+W LGPVSL N+D +KA RG +E E L
Sbjct: 388 SVFNSFYKLESEYYDHYKKVMGTKSWGLGPVSLWANQDDSDKAARGYARKEEGAKEEGWL 567
Query: 268 KWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLP 327
KWL SK SV+YV FGSM F +QL EIA LE S FIWVVRK+ ++E +
Sbjct: 568 KWLNSKPDGSVLYVSFGSMNKFPYSQLVEIAHALENSGHNFIWVVRKNEENEEGGV--FL 741
Query: 328 EGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYG 387
E FE++++ SGKG +I GWAPQ++IL++ ++GG V+HCGWN+ +E V+ GLP VTWP++
Sbjct: 742 EEFEKKMKESGKGYLIWGWAPQLLILENHAIGGLVSHCGWNTVVESVNVGLPTVTWPLFA 921
Query: 388 EQFYNAKFLSDIVKIGVGVGVQTWIGMG--GGEPVKKDVIEKAVRRIMVGDEAE-EMRSR 444
E F+N K + D++KIGV VG + W G E VK++ I A+R +M G E E MR R
Sbjct: 922 EHFFNEKLVVDVLKIGVPVGAKEWRNWNEFGSEVVKREDIGNAIRLMMEGGEEEVAMRKR 1101
Query: 445 AKEFGKMARRAVEVGGSSYNDFSNLIEDLKS 475
KE A++A++VGGSSYN+ LI++L+S
Sbjct: 1102VKELSVEAKKAIKVGGSSYNNMVELIQELRS 1194
>TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(26%)
Length = 1248
Score = 273 bits (698), Expect(2) = 7e-75
Identities = 152/377 (40%), Positives = 214/377 (56%), Gaps = 8/377 (2%)
Frame = +3
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFP 67
L + F P+ A+GH++P D+A +F+SRG VTI+TT N +++T+ A + + T++FP
Sbjct: 99 LKLHFIPYPASGHMMPLCDIATLFASRGQHVTIITTPSNAQSLTKTLSSAALRLHTVEFP 278
Query: 68 SPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWSTD 127
++ LP+G E+ S P K +LL E + +E+ PDC++AD F W+ D
Sbjct: 279 Y-QQVDLPKGVESMTSTTDPITTWKIHNGAMLLNEAVGDFVEKNPPDCIIADSAFSWAND 455
Query: 128 SAAKFNIPRIVFHGLGFFPLCVLACTRQYK---PQDKVSSYTEPFVVPNLPGEITLTKMQ 184
A K IP + F+G F + + R S + +VVPNL + +T
Sbjct: 456 LAHKLQIPNLTFNGSSLFAVSIFHSLRTNNLLHTDADADSDSSSYVVPNLHHD-NITLCS 632
Query: 185 LPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELE-PVYADHYRNELGRKAWHLGPVS 243
P KV + + ++ +KS G I N F EL+ HY G KAWHLGP S
Sbjct: 633 KPP-----KVLSMFIGMMLDTVLKSTGYIINNFVELDGEECVKHYEKTTGHKAWHLGPTS 797
Query: 244 LCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEA 303
+ +EKA +G ++ + EHECL WL+S+ NSV+Y+CFGS+ FSD QL EIA +EA
Sbjct: 798 FIRKTVQEKAEKGNKSDVSEHECLNWLKSQRVNSVVYICFGSINHFSDKQLYEIACAVEA 977
Query: 304 SEVPFIWVV----RKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVG 359
S PFIWVV K + E E +WLP+GFEER G KG IIRGWAPQV+IL + +VG
Sbjct: 978 SGHPFIWVVPEKKGKEDEIEEEKEKWLPKGFEERNIGK-KGFIIRGWAPQVLILSNPAVG 1154
Query: 360 GFVTHCGWNSTLEGVSA 376
GF+THCG G+ A
Sbjct: 1155GFLTHCGGKLNCRGLWA 1205
Score = 26.2 bits (56), Expect(2) = 7e-75
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = +2
Query: 374 VSAGLPMVTWPMYGE 388
V AG+PM+TWP + +
Sbjct: 1199 VGAGVPMITWPCHAD 1243
>TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (38%)
Length = 1081
Score = 277 bits (708), Expect = 7e-75
Identities = 142/256 (55%), Positives = 178/256 (69%), Gaps = 8/256 (3%)
Frame = +2
Query: 227 HYRNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSM 286
HY G K WHLGP SL + +EK+ RG E +++ HE L WL S+ NSV+Y+CFGS+
Sbjct: 5 HYEKATGHKVWHLGPTSLIRKTAQEKSERGNEGAVNVHESLSWLDSERVNSVLYICFGSI 184
Query: 287 TVFSDAQLKEIAMGLEASEVPFIWVVR----KSAKSEGENLEWLPEGFEERIEGSGKGLI 342
FSD QL E+A +EAS PFIWVV K +SE E +WLP+GFEER G KGLI
Sbjct: 185 NYFSDKQLYEMACAIEASGHPFIWVVPEKKGKEDESEEEKEKWLPKGFEERNIGK-KGLI 361
Query: 343 IRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKI 402
IRGWAPQV IL H +VGGF+THCG NST+E VSAG+PM+TWP++G+QFYN K ++ I
Sbjct: 362 IRGWAPQVKILSHPAVGGFMTHCGGNSTVEAVSAGVPMITWPVHGDQFYNEKLITQFRGI 541
Query: 403 GVGVGVQTWIGMGGGEP---VKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEV 458
GV VG W G E V +D IEKAVRR+M GDEAE +R RA+EFG+ A +A++
Sbjct: 542 GVEVGATEWCTSGVAERKKLVSRDSIEKAVRRLMDGGDEAENIRLRAREFGEKAIQAIQE 721
Query: 459 GGSSYNDFSNLIEDLK 474
GGSSYN+ LI++LK
Sbjct: 722 GGSSYNNLLALIDELK 769
>TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose: flavonoid
7-O-glucosyltransferase {Scutellaria baicalensis},
partial (29%)
Length = 843
Score = 257 bits (657), Expect = 6e-69
Identities = 126/228 (55%), Positives = 164/228 (71%), Gaps = 8/228 (3%)
Frame = +1
Query: 255 RGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRK 314
RG E+ + HECL WL SK+ NSV+Y+CFGS FSD QL EIA G+EAS FIWVV +
Sbjct: 4 RGEESVVSVHECLSWLNSKQDNSVVYICFGSQCHFSDKQLYEIACGIEASSHEFIWVVPE 183
Query: 315 SAKSEGENLE----WLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNST 370
++E +N E WLP+GFEERI G K +II+GWAPQVMIL H +VG F+THCGWNST
Sbjct: 184 KKRTENDNEEEKEKWLPKGFEERIIGKKKAMIIKGWAPQVMILSHTAVGAFMTHCGWNST 363
Query: 371 LEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVK---KDVIEK 427
+E VSAG+PM+TWPM+GEQFYN K ++ + IGV VG W G GE K +D IEK
Sbjct: 364 VEAVSAGVPMITWPMHGEQFYNEKLITQVHGIGVEVGATEWSTTGIGEREKVVWRDNIEK 543
Query: 428 AVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLK 474
V+R+M GDEAE++R A+EFG+ A+ A++ GGSS+++ + ++ LK
Sbjct: 544 VVKRLMDSGDEAEKIRQHAREFGEKAKHAIKEGGSSHSNLTAVVNYLK 687
>TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(19%)
Length = 1118
Score = 244 bits (622), Expect = 7e-65
Identities = 138/347 (39%), Positives = 198/347 (56%), Gaps = 11/347 (3%)
Frame = +3
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK---INIKTI 64
L + F PFLA+GH+IP D+A +F+S G +VT++TT N +++++ A + + T+
Sbjct: 105 LKLHFIPFLASGHMIPLFDIATMFASHGHQVTVITTPSNAQSLTKSLSSAASFFLRLHTV 284
Query: 65 KFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPW 124
FPS E+ LP+G E+ S K + +LL P+E+ +E++ PDC+++D +PW
Sbjct: 285 DFPS-EQVDLPKGIESMSSTTDSITSWKIHRGAMLLHGPIENFMEKDPPDCIISDSTYPW 461
Query: 125 STDSAAKFNIPRIVFHGLGFFPLCV---LACTRQYKPQDKVSSYTEPFVVPNLPGEITLT 181
+ D A K IP + F+GL F + + L S + F+VPN P ITL+
Sbjct: 462 ANDLAHKLQIPNLTFNGLSLFTISLVESLIRNNLLHSDTNSDSDSSSFLVPNFPHRITLS 641
Query: 182 KMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYA-DHYRNELGRKAWHLG 240
+ + KV +K L+ E+ +KS +I N F EL+ HY GRK WHLG
Sbjct: 642 E-------KPPKVLSKFLKMMLETVLKSKALIINNFAELDGEECIQHYEKTTGRKVWHLG 800
Query: 241 PVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMG 300
P SL R +EKA RG E ++ HECL WL S+ N+V+Y+CFGS+ SD QL E+A
Sbjct: 801 PTSLIRRTIQEKAERGNEGEVNMHECLSWLNSQRVNAVLYICFGSINYLSDKQLYEMACA 980
Query: 301 LEASEVPFIWVV----RKSAKSEGENLEWLPEGFEERIEGSGKGLII 343
+E S PFIWVV K +SE E +WLP+GFEER S GLII
Sbjct: 981 IETSGHPFIWVVPEKKGKEDESEEEKEKWLPKGFEER-NISKMGLII 1118
>TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (42%)
Length = 1071
Score = 243 bits (621), Expect = 9e-65
Identities = 125/222 (56%), Positives = 156/222 (69%), Gaps = 8/222 (3%)
Frame = +2
Query: 261 IDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVR----KSA 316
+ HECL WL SKE NSV+Y+CFGS++ FSD QL EIA G+E S F+WVV K
Sbjct: 2 VSVHECLSWLDSKEDNSVLYICFGSISHFSDKQLYEIASGIENSGHKFVWVVPEKKGKED 181
Query: 317 KSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSA 376
+SE + +WLP+GFEER + KG II+GWAPQ MIL H VG F+THCGWNS +E +SA
Sbjct: 182 ESEEQKEKWLPKGFEERNILNKKGFIIKGWAPQAMILSHTVVGAFMTHCGWNSIVEAISA 361
Query: 377 GLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEP---VKKDVIEKAVRRIM 433
G+PM+TWP++GEQFYN K ++ + +IGV VG W G E V + IEKAVRR+M
Sbjct: 362 GIPMITWPVHGEQFYNEKLITVVQRIGVEVGATEWSLHGFQEKEKVVSRHSIEKAVRRLM 541
Query: 434 -VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLK 474
GDEA+E+R RA+EFGK A AVE GGSS+N+ LI DLK
Sbjct: 542 DDGDEAKEIRQRAQEFGKKATHAVEEGGSSHNNLLALINDLK 667
>BQ123034 weakly similar to GP|14334982|gb putative glucosyltransferase
{Arabidopsis thaliana}, partial (24%)
Length = 765
Score = 214 bits (544), Expect = 8e-56
Identities = 113/221 (51%), Positives = 150/221 (67%), Gaps = 5/221 (2%)
Frame = +2
Query: 260 SIDEH-ECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKS 318
S+ +H E L WL SKE SV+YV FGS T AQL EI GLE S FIWV+++
Sbjct: 56 SLGKHTELLNWLNSKENESVLYVSFGSFTRLPYAQLVEIVHGLENSGHNFIWVIKRD--D 229
Query: 319 EGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGL 378
E+ E + FEERI+ S KG II WAPQ++ILDH + GG VTHCGWNSTLE ++AGL
Sbjct: 230 TDEDGEGFLQEFEERIKESSKGYIIWDWAPQLLILDHPATGGIVTHCGWNSTLESLNAGL 409
Query: 379 PMVTWPMYGEQFYNAKFLSDIVKIGVGVGV---QTWIGMGGGEPVKKDVIEKAVRRIM-V 434
PM+TWP++ EQFYN K L D++KIGV VG + W+ + + V+++ IEK V+ +M
Sbjct: 410 PMITWPIFAEQFYNEKLLVDVLKIGVPVGAKENKLWLDISVEKVVRREEIEKTVKILMGS 589
Query: 435 GDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKS 475
G E++EMR RAK+ + A+R +E GG SYN+ LI++LKS
Sbjct: 590 GQESKEMRMRAKKLSEAAKRTIEEGGDSYNNLIQLIDELKS 712
>BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosyltransferase
{Vigna mungo}, partial (21%)
Length = 646
Score = 205 bits (521), Expect = 4e-53
Identities = 98/175 (56%), Positives = 123/175 (70%), Gaps = 4/175 (2%)
Frame = +1
Query: 224 YADHYRNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCF 283
Y +Y + G KAWHLGP SL + +EKA RG+E+++ ECL WL SK NSV+Y+ F
Sbjct: 97 YIKYYVSSTGHKAWHLGPASLIRKTVQEKAERGQESAVSVQECLSWLNSKRHNSVLYISF 276
Query: 284 GSMTVFSDAQLKEIAMGLEASEVPFIWVV----RKSAKSEGENLEWLPEGFEERIEGSGK 339
GS+ + D QL EIA +EAS FIWV+ K +SE E +WLP+GFEER G K
Sbjct: 277 GSLCRYQDKQLYEIACAIEASGHDFIWVIPLNNGKEDESEEEKQKWLPKGFEERNIGK-K 453
Query: 340 GLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAK 394
GLIIRGWAPQV+IL H +VG F+THCGWNST+E V AG+PM+TWP +GEQF + K
Sbjct: 454 GLIIRGWAPQVLILSHPAVGAFMTHCGWNSTVEAVGAGVPMITWPSHGEQFLHRK 618
>TC81362 similar to GP|19911201|dbj|BAB86927. glucosyltransferase-9 {Vigna
angularis}, partial (70%)
Length = 1347
Score = 202 bits (514), Expect = 2e-52
Identities = 112/307 (36%), Positives = 176/307 (56%), Gaps = 10/307 (3%)
Frame = +1
Query: 9 HIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA-----KINIKT 63
H + FP +A GHIIP +D+A++ + RG+ VTI TT N + + +A +I I T
Sbjct: 106 HFVLFPLIAQGHIIPMIDIAKLLAQRGVIVTIFTTPKNASRFTSVLSRAVSSGLQIKIVT 285
Query: 64 IKFPSPEETGLPEGCENSESA-LAPDKFIKF--MKSTLLLREPLEHVLEQ--EKPDCLVA 118
+ FPS ++ GLP+GCEN + ++ D +K+ + LL++ E + ++ KP C+++
Sbjct: 286 LNFPS-KQVGLPDGCENFDMVNISKDMNMKYNLFHAVSLLQKEGEDLFDKLSPKPSCIIS 462
Query: 119 DMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEI 178
D W++ A K +IPRI FHG F L + + ++S TE F +P +P +I
Sbjct: 463 DFCITWTSQIAEKHHIPRISFHGFCCFTLHCMFKVHTSNILESINSETEFFSIPGIPDKI 642
Query: 179 TLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWH 238
+TK Q+P + +K+ E+ E+E+KS+GVI N+F ELE Y + Y+ K W
Sbjct: 643 QVTKEQIPGTVKEEKM-KGFAEKMQEAEMKSYGVIINSFEELEKEYVNDYKKVRNDKVWC 819
Query: 239 LGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIA 298
+GPV+LCN+D +KA RG ASI EH CL +L +P SV+YVC G F+ I
Sbjct: 820 VGPVALCNKDGLDKAQRGNIASISEHNCLNFLDLHKPKSVVYVCLGKFMQFNSFTTY*IG 999
Query: 299 MGLEASE 305
G+ +++
Sbjct: 1000FGITSNK 1020
Score = 150 bits (378), Expect = 1e-36
Identities = 66/119 (55%), Positives = 94/119 (78%), Gaps = 1/119 (0%)
Frame = +2
Query: 292 AQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLP-EGFEERIEGSGKGLIIRGWAPQV 350
+QL E+A+GL+A+++PFIWV+R+ E +W+ E FEER G+GLIIRGWAPQ+
Sbjct: 980 SQLIELALGLQATKIPFIWVIREGIYKSEELEKWISDEKFEER--NKGRGLIIRGWAPQM 1153
Query: 351 MILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQ 409
+IL H S+GGF+THCGWNSTLEG+S G+PMVTWP++ +QF N K ++ +++IGV +GV+
Sbjct: 1154 VILSHSSIGGFLTHCGWNSTLEGISFGVPMVTWPLFADQFLNEKLVTQVLRIGVSLGVE 1330
>CA918173 weakly similar to GP|11034537|dbj putative glucosyl transferase
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 769
Score = 198 bits (504), Expect = 3e-51
Identities = 107/229 (46%), Positives = 147/229 (63%), Gaps = 3/229 (1%)
Frame = +2
Query: 171 VPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRN 230
+P LP I +T++ L + + T + E ES +S+G + N+F+ELE Y ++
Sbjct: 8 IPCLPHTIEMTRLHLHNWERENNAMTAIFEPMYESAERSYGSLYNSFHELESDYEKLFKT 187
Query: 231 ELGRKAWHLGPVSL-CNRDTEEKACRGR-EASIDEH-ECLKWLQSKEPNSVIYVCFGSMT 287
+G K+W +GPVS N+D E KA RG E S+ +H E L WL SKE SV+YV FGS+
Sbjct: 188 TIGIKSWSVGPVSAWANKDDERKANRGHIEKSLGKHTELLNWLNSKENESVLYVSFGSLI 367
Query: 288 VFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWA 347
AQL EIA GLE S FIWV++ + +GE +L E FE+R++ S KG II WA
Sbjct: 368 RLPHAQLVEIAHGLENSGHNFIWVIKNNKDEDGEG--FLQE-FEKRMKESNKGYIIWDWA 538
Query: 348 PQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFL 396
PQ++IL++ ++GG VTHCGWNSTLE V+AGLPM+TWP++ E F K L
Sbjct: 539 PQLLILEYPAIGGIVTHCGWNSTLESVNAGLPMITWPVFAEDFTMRKCL 685
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 197 bits (500), Expect = 1e-50
Identities = 146/477 (30%), Positives = 224/477 (46%), Gaps = 17/477 (3%)
Frame = +3
Query: 10 IIFFPFLANGHIIPCVDLARVF----SSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIK 65
I P H+IP V+ A++ + + I T P + + N+ I
Sbjct: 66 IAMVPSPGLSHLIPLVEFAKLLLQNHNEYHITFLIPTLGPLTPSMQSILNTLPPNMNFIV 245
Query: 66 FPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWS 125
P LP N + A +K S L E ++ +L + + LV MF +
Sbjct: 246 LPQVNIEDLPH---NLDPATQMKLIVKH--SIPFLYEEVKSLLSKTRLVALVFSMFSTDA 410
Query: 126 TDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPG-EITLTKMQ 184
D A FN+ +F G + + N+PG I L +
Sbjct: 411 HDVAKHFNLLSYLFFSSGAVLFSLFLTIPNLDEAASTQFLGSSYETVNIPGFSIPLHIKE 590
Query: 185 LPQ---LPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGP 241
LP + + +L+ + + GVI NTF +LEP ++ + +GP
Sbjct: 591 LPDPFICERSSDAYKSILDVCQKLSLFD-GVIMNTFTDLEPEVIRVLQDREKPSVYPVGP 767
Query: 242 VSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGL 301
+ + E CL+WL++++P+SV++V FGS S QL EIA GL
Sbjct: 768 MIRNESNNEANMSM----------CLRWLENQQPSSVLFVSFGSGGTLSQDQLNEIAFGL 917
Query: 302 EASEVPFIWVVRKSAKSE------GEN---LEWLPEGFEERIEGSGKGLIIRGWAPQVMI 352
E S F+WVVR +K+ G+N LE+LP GF ER + +G L++ WAPQV I
Sbjct: 918 ELSGHKFLWVVRAPSKNSSSAYFSGQNNDPLEYLPNGFLERTKENG--LVVASWAPQVEI 1091
Query: 353 LDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWI 412
L H S+GGF++HCGW+STLE V G+P++ WP++ EQ NAK L+D++K+ V V
Sbjct: 1092LGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVAVRPKVDDET 1271
Query: 413 GMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNL 469
G+ +K++ + KA++RIM GDE+ E+R + KE A + GSS S+L
Sbjct: 1272GI-----IKQEEVAKAIKRIMKGDESFEIRKKIKELSVGAATVLSEHGSSRKALSSL 1427
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 196 bits (498), Expect = 2e-50
Identities = 139/485 (28%), Positives = 232/485 (47%), Gaps = 15/485 (3%)
Frame = +2
Query: 4 ENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKT 63
+N H + P+ A GH+ P + ++ +G+K+T++T +IS N+ +
Sbjct: 29 KNHAPHCLILPYPAQGHMNPMIQFSKRLIEKGVKITLITVTSFWKVISNK------NLTS 190
Query: 64 IKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKP-DCLVADMFF 122
I S + G EG + +L K + + L E L + E P +C++ D F
Sbjct: 191 IDVESISD-GYDEGGLLAAESLEDYKETFWKVGSQTLSELLHKLSSSENPPNCVIFDAFL 367
Query: 123 PWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKV-------SSYTEPFVVPNLP 175
PW D F + G+ FF + Y +K+ S Y P + P
Sbjct: 368 PWVLDVGKSFGLV-----GVAFFTQSCSVNSVYYHTHEKLIELPLTQSEYLLPGLPKLAP 532
Query: 176 GEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRK 235
G++ + P + + +++ ++AN+ YELEP D K
Sbjct: 533 GDLPSFLYKYGSYPGYFDIVVNQFANIGKAD----WILANSIYELEPEVVDWLV-----K 685
Query: 236 AWHLG------PVSLCNRDTEEKACRGREASIDEHE-CLKWLQSKEPNSVIYVCFGSMTV 288
W L P L ++ ++ G S E C+KWL K SV+Y FGSM
Sbjct: 686 IWPLKTIGPSVPSMLLDKRLKDDKEYGVSLSDPNTEFCIKWLNDKPKGSVVYASFGSMAG 865
Query: 289 FSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAP 348
S+ Q +E+A+GL+ SE F+WVVR+ +S+ LP+GF +E S KGLI+ W P
Sbjct: 866 LSEEQTQELALGLKDSESYFLWVVRECDQSK------LPKGF---VESSKKGLIVT-WCP 1015
Query: 349 QVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGV 408
Q+++L HE++G FVTHCGWNSTLE +S G+P++ P++ +Q NAK ++D+ K+GV
Sbjct: 1016QLLVLTHEALGCFVTHCGWNSTLEALSIGVPLIAMPLWTDQVTNAKLIADVWKMGVRAVA 1195
Query: 409 QTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
E V+ + I+ ++ I+ ++ E++ A ++ +A+ +V+ GG S +
Sbjct: 1196DE------KEIVRSETIKNCIKEIIETEKGNEIKKNALKWKNLAKSSVDEGGRSDKNIEE 1357
Query: 469 LIEDL 473
+ L
Sbjct: 1358FVAAL 1372
>TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (31%)
Length = 3139
Score = 193 bits (490), Expect = 1e-49
Identities = 149/486 (30%), Positives = 226/486 (45%), Gaps = 23/486 (4%)
Frame = -1
Query: 7 ELHII-------FFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKI 59
ELHI+ P H+IP V+ A++ + I +P + +
Sbjct: 2800 ELHIMENKTCIAMVPSPGLSHLIPQVEFAKLLLQHHNESHITFL---IPTLGPLTPSMQS 2630
Query: 60 NIKTIKFPSPEETGLPE-GCENSESALAPDKFIKFM--KSTLLLREPLEHVLEQEKPDCL 116
+ T+ P+ T LP+ E+ L P +K + S L E ++ +L + L
Sbjct: 2629 ILNTLP-PNMNFTVLPQVNIEDLPHNLEPSTQMKLIVKHSIPFLHEEVKSLLSKTNLVAL 2453
Query: 117 VADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPG 176
V MF + D A FN+ +F G + + N+PG
Sbjct: 2452 VCSMFSTDAHDVAKHFNLLSYLFFSSGAVLFSFFLTLPNLDDAASTQFLGSSYEMVNVPG 2273
Query: 177 -EITLTKMQLPQ---LPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNEL 232
I +LP + + +L+ +S + GVI NTF LE ++
Sbjct: 2272 FSIPFHVKELPDPFNCERSSDTYKSILDVCQKSSLFD-GVIINTFSNLELEAVRVLQDRE 2096
Query: 233 GRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDA 292
+ +GP+ + N E + CL+WL+++ P+SVI+V FGS S
Sbjct: 2095 KPSVFPVGPI-IRNESNNEA---------NMSVCLRWLENQPPSSVIFVSFGSGGTLSQD 1946
Query: 293 QLKEIAMGLEASEVPFIWVVRKSAK---------SEGENLEWLPEGFEERIEGSGKGLII 343
QL E+A GLE S F+WVVR +K E LE+LP GF ER + KGL++
Sbjct: 1945 QLNELAFGLELSGHKFLWVVRAPSKHSSSAYFNGQNNEPLEYLPNGFVERTKE--KGLVV 1772
Query: 344 RGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIG 403
WAPQV IL H S+GGF++HCGW+STLE V G+P++ WP++ EQ NAK L+D++K+
Sbjct: 1771 TSWAPQVEILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLKVA 1592
Query: 404 VGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSY 463
V V G+ +K++ + KA++RIM GDE+ E+R + KE A + GSS
Sbjct: 1591 VRPKVDGETGI-----IKREEVSKALKRIMEGDESFEIRKKIKELSVSAATVLSEHGSSR 1427
Query: 464 NDFSNL 469
S L
Sbjct: 1426 KALSTL 1409
>TC85817 weakly similar to PIR|B85014|B85014 probable flavonol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (83%)
Length = 1736
Score = 192 bits (487), Expect = 3e-49
Identities = 149/487 (30%), Positives = 237/487 (48%), Gaps = 27/487 (5%)
Frame = +3
Query: 10 IIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFPSP 69
++ P GH+IP ++ A+ ++ I+ +L + T G KT+ P
Sbjct: 162 VVMLPSPGMGHLIPMIEFAK-------RIIILNQNLQITFFIPTEGPPSKAQKTVLQSLP 320
Query: 70 E---ETGLPEGCENSESALAPDKFIK------FMKSTLLLREPLEHVLEQEKPDCLVADM 120
+ T LP S S L P+ I+ ++S LR+ + E +V D+
Sbjct: 321 KFISHTFLPPV---SFSDLPPNSGIETIISLTVLRSLPSLRQNFNTLSETHTITAVVVDL 491
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQD--KVSSYTEPFVVPN---LP 175
F + D A +FN+P+ VF+ L + + + + TEP +P +
Sbjct: 492 FGTDAFDVAREFNVPKYVFYPSTAMALSLFLYLPRLDEEVHCEFRELTEPVKIPGCIPIH 671
Query: 176 GEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEP-VYADHYRNELGR 234
G+ L +Q + + VF + E++ G+I N+F ELEP + + E G+
Sbjct: 672 GKYLLDPLQDRKNDAYQSVFRNA-KRYREAD----GLIENSFLELEPGPIKELLKEEPGK 836
Query: 235 -KAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQ 293
K + +GP L R+ E E LKWL ++ SV++V FGS S Q
Sbjct: 837 PKFYPVGP--LVKREVEVGQIGPNS------ESLKWLDNQPHGSVLFVSFGSGGTLSSKQ 992
Query: 294 LKEIAMGLEASEVPFIWVVRK-----------SAKSEGENLEWLPEGFEERIEGSGKGLI 342
+ E+A+GLE SE F+WVVR SA+++ + ++LP GF ER +G +GL+
Sbjct: 993 IVELALGLEMSEQRFLWVVRSPNDKVANASYFSAETDSDPFDFLPNGFLERTKG--RGLV 1166
Query: 343 IRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKI 402
+ WAPQ +L H S GGF+THCGWNS LE V G+P+V WP+Y EQ NA L++ VK+
Sbjct: 1167VSSWAPQPQVLAHGSTGGFLTHCGWNSVLESVVNGVPLVVWPLYAEQKMNAVMLTEDVKV 1346
Query: 403 GVGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSS 462
G+ V G V++ I V+ +M G+E +++R + K+ + A + + G+S
Sbjct: 1347GLRPNV------GENGLVERLEIASVVKCLMEGEEGKKLRYQMKDLKEAASKTLGENGTS 1508
Query: 463 YNDFSNL 469
N SNL
Sbjct: 1509TNHISNL 1529
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 191 bits (486), Expect = 4e-49
Identities = 158/505 (31%), Positives = 239/505 (47%), Gaps = 30/505 (5%)
Frame = +3
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLAR--VFSSRGLKVTIVTTHLNVPLISRTIGKAK 58
+T + +HI P H++P V+ ++ V + VT + L P S K
Sbjct: 12 LTKMAKTIHIAVIPSPGFSHLVPIVEFSKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLEK 191
Query: 59 I--NIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLL--LREPLEHVLEQEKPD 114
I NI +I P + LP+G + P I+ + L + + L+ + +
Sbjct: 192 IPPNINSIFLPPINKQDLPQG-------VYPAILIQQTVTLSLPSIHQALKSLSSKAPLV 350
Query: 115 CLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQY-----KPQDKVSSYTEPF 169
++AD F + D A +FN L +F A + K ++VS +
Sbjct: 351 AIIADSFAFEALDFAKEFN-------SLSYFYFPTSANVLSFILHLPKLDEEVSCEFKDL 509
Query: 170 VVP-NLPGEITLTKMQLPQLPQHDK---VFTKLLEESNESEVKSF----GVIANTFYELE 221
P L G + + + LP P D+ + L+ + KSF G++ N+FYELE
Sbjct: 510 QEPIKLQGCVPINGIDLPT-PTKDRSSEAYRMFLQRA-----KSFYFVDGILINSFYELE 671
Query: 222 PVYADHYRNE-LGRKAWH-LGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVI 279
+ + + G ++ +GP++ + DEHECLKWL+++ NSV+
Sbjct: 672 SSAVEALKQKGYGNISYFPVGPITQIGSSNNDVVG-------DEHECLKWLKNQPQNSVL 830
Query: 280 YVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKS------EGEN---LEWLPEGF 330
YV FGS S Q+ EIA GLE S FIW VR + S E N L++LPEGF
Sbjct: 831 YVSFGSGGTLSQRQINEIAFGLELSGQRFIWGVRAPSDSVNAAYLESTNEDPLKFLPEGF 1010
Query: 331 EERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQF 390
+ER + KG I+ WAPQV IL H SVGGF++HCGWNS LE + G+P+V WP++ EQ
Sbjct: 1011QERTKE--KGFILPSWAPQVEILKHSSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQA 1184
Query: 391 YNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGK 450
NA L D +K+ + + + E V+KD ++ +M G+E + MR R K
Sbjct: 1185MNAVMLCDGLKVAIRLKFE------DDEIVEKDKTANVIKCLMEGEEGKTMRDRMKSLKD 1346
Query: 451 MARRAVEVGGSSYNDFSNLIEDLKS 475
A AV+ GSS + S L +S
Sbjct: 1347YAVNAVKDEGSSIQNLSQLASQWES 1421
>TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (27%)
Length = 1669
Score = 191 bits (485), Expect = 5e-49
Identities = 144/494 (29%), Positives = 233/494 (47%), Gaps = 22/494 (4%)
Frame = +2
Query: 2 TNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINI 61
T E+H++ F A GHI P + L + ++GL VT+ TT L + R +
Sbjct: 53 TEIKPEIHVLLVAFSAQGHINPLLRLGKSLLTKGLNVTLATTEL---VYHRVFKSTTTDT 223
Query: 62 KTIKFPSPEETG------LPEGCENSESALAPDKFIKFMKS------TLLLREPLEHVLE 109
T+ P+ T +G + S+ +PD++++ + T L+ +
Sbjct: 224 DTV--PTSYSTNGINVLFFSDGFDISQGHKSPDEYMELIAKFGPISLTNLINNNFINN-P 394
Query: 110 QEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACT------RQYKPQDKVS 163
+K C++ + F PW + A + NIP L + CT R Y ++
Sbjct: 395 SKKLACIINNPFVPWVANVAYELNIPCAC--------LWIQPCTLYSIYYRFYNELNQFP 550
Query: 164 SYTEPFVVPNLPGEITLTKMQLPQ--LPQHD-KVFTKLLEESNESEVKSFGVIANTFYEL 220
+ P + LPG L LP LP + K + +L E + K V+AN+FYEL
Sbjct: 551 TLENPEIDVELPGLPLLKPQDLPSFILPSNPIKTMSDVLAEMFQDMKKLKWVLANSFYEL 730
Query: 221 EPVYADHYRNELG-RKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVI 279
E D L P SL +D ++ G E + C++WL K P+SVI
Sbjct: 731 EKDVIDSMAEIFPITPVGPLVPPSLLGQDQKQDV--GIEMWTPQDSCMEWLNQKLPSSVI 904
Query: 280 YVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGK 339
Y+ FGS+ S+ Q++ IA L+ S F+WV++ + G L E F E E K
Sbjct: 905 YISFGSLIFLSEKQMQSIASALKNSNKYFLWVMKSN--DIGAEKVQLSEKFLE--ETKEK 1072
Query: 340 GLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDI 399
GL++ W PQ +L H ++ F+THCGWNSTLE ++AG+PM+ +P + +Q NA +SD+
Sbjct: 1073GLVVT-WCPQTKVLVHPAIACFLTHCGWNSTLEAITAGVPMIGYPQWTDQPTNATLVSDV 1249
Query: 400 VKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVG 459
++G+ + V+ D +E+A+ I+ G+ +E + A E AR AV G
Sbjct: 1250FRMGIR------LKQDNDGFVESDEVERAIEEIVGGERSEVFKKNALELKHAAREAVADG 1411
Query: 460 GSSYNDFSNLIEDL 473
GSS + ++++
Sbjct: 1412GSSDRNIQRFVDEI 1453
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,076,722
Number of Sequences: 36976
Number of extensions: 221621
Number of successful extensions: 1432
Number of sequences better than 10.0: 196
Number of HSP's better than 10.0 without gapping: 1209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1231
length of query: 478
length of database: 9,014,727
effective HSP length: 100
effective length of query: 378
effective length of database: 5,317,127
effective search space: 2009874006
effective search space used: 2009874006
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136449.6


