

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.13 + phase: 0
(373 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
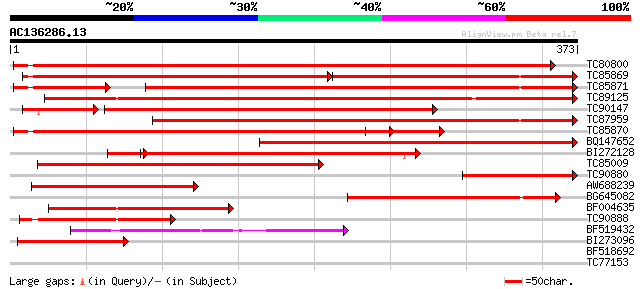
Score E
Sequences producing significant alignments: (bits) Value
TC80800 similar to GP|6606532|gb|AAF19195.1| pectate lyase 1 {Mu... 446 e-126
TC85869 homologue to GP|14531296|gb|AAK66161.1 pectate lyase {Fr... 271 e-120
TC85871 similar to GP|7547009|gb|AAF63756.1| pectate lyase {Viti... 350 e-114
TC89125 similar to PIR|H86253|H86253 hypothetical protein [impor... 373 e-104
TC90147 similar to GP|21537059|gb|AAM61400.1 pectate lyase-like ... 290 5e-93
TC87959 similar to SP|P24396|9612_LYCES Style development-specif... 323 9e-89
TC85870 similar to GP|18087517|gb|AAL58893.1 At1g67750/F12A21_12... 322 1e-88
BQ147652 homologue to PIR|T09524|T09 probable pectate lyase (EC ... 275 3e-74
BI272128 similar to PIR|T09524|T09 probable pectate lyase (EC 4.... 250 4e-71
TC85009 similar to GP|14531294|gb|AAK66160.1 pectate lyase B {Fr... 238 2e-63
TC90880 similar to GP|15027941|gb|AAK76501.1 unknown protein {Ar... 159 2e-39
AW688239 similar to GP|21537059|gb pectate lyase-like protein {A... 147 5e-36
BG645082 similar to GP|20466073|gb At5g04300/At5g04300 {Arabidop... 147 5e-36
BF004635 similar to PIR|H86253|H86 hypothetical protein [importe... 135 2e-32
TC90888 similar to GP|14289169|dbj|BAB59066. pectate lyase {Sali... 128 3e-30
BF519432 similar to GP|6682233|gb| putative pectate lyase {Arabi... 103 1e-22
BI273096 similar to GP|18087517|gb At1g67750/F12A21_12 {Arabidop... 100 9e-22
BF518692 similar to PIR|S51509|S515 pectase lyase - Aspergillus ... 32 0.52
TC77153 homologue to GP|21902507|gb|AAM78552.1 ribosomal protein... 27 9.7
>TC80800 similar to GP|6606532|gb|AAF19195.1| pectate lyase 1 {Musa
acuminata}, partial (86%)
Length = 1216
Score = 446 bits (1148), Expect = e-126
Identities = 212/357 (59%), Positives = 258/357 (71%)
Frame = +1
Query: 3 IGGINGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPI 62
I G RRNL G SC + NPID CWRCDPNW NR++LADC GFG+ GGK+G I
Sbjct: 139 INGSLARRNL---GYLSCGSGNPIDDCWRCDPNWEKNRQRLADCAIGFGKNAIGGKNGKI 309
Query: 63 YVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADV 122
YVVTD D D V P+PGTLR AV ++ PLWIIFAR M I+L +ELIM KTIDGRGA V
Sbjct: 310 YVVTDSGDDDPVTPKPGTLRFAVIQDEPLWIIFARDMVIQLKEELIMNSFKTIDGRGASV 489
Query: 123 TIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHI 182
IA G ITIQ++ NVIIHGI I+D G +VRDS HYG RT+SDGDG+SIFG SH+
Sbjct: 490 HIAGGPCITIQYVTNVIIHGIHIHDCKQGGNAMVRDSPGHYGWRTVSDGDGVSIFGGSHV 669
Query: 183 WIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNR 242
W+DH S+ NC DGLIDAI GSTAITISN++ T H++VML G SDTY D+ MQ+T+ FN
Sbjct: 670 WVDHCSLSNCNDGLIDAIHGSTAITISNNYMTHHDKVMLLGHSDTYTQDKNMQVTIAFNH 849
Query: 243 FGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITK 302
FG+ L+QRMPRCR G+ HV+NN Y WEMYAIGG+ +PTI S+GN+F+AP++ +KE+TK
Sbjct: 850 FGEGLVQRMPRCRHGYFHVVNNDYTHWEMYAIGGSANPTINSQGNRFVAPDDRFSKEVTK 1029
Query: 303 RTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLT 359
PE EWK W WRS DL +NGAFF SGA + ++ + A+P S VG +T
Sbjct: 1030HEDAPEGEWKGWNWRSEGDLLINGAFFTPSGAGGASSSYARXSSLSARPSSLVGTIT 1200
>TC85869 homologue to GP|14531296|gb|AAK66161.1 pectate lyase {Fragaria x
ananassa}, complete
Length = 2263
Score = 271 bits (694), Expect(2) = e-120
Identities = 130/204 (63%), Positives = 153/204 (74%)
Frame = +3
Query: 9 RRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
RRNL G SC NPID CWRCD NW NRK+LADC GFGR GG+DG YVVTDP
Sbjct: 423 RRNL---GFFSCGTGNPIDDCWRCDRNWQQNRKRLADCGIGFGRNAIGGRDGKYYVVTDP 593
Query: 69 SDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGA 128
D D VNPRPGTLRHAV ++ PLWI+F R M I+ QELI+ KTIDGRGA+V IANG
Sbjct: 594 RDDDPVNPRPGTLRHAVIQDRPLWIVFKRDMVIQFKQELIVNSFKTIDGRGANVHIANGG 773
Query: 129 GITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVS 188
ITIQ++ NVIIHG+ I+D +VR SE H+G RT++DGD +SIFGSSHIW+DH S
Sbjct: 774 CITIQYVTNVIIHGLHIHDCKPTGNAMVRSSETHFGWRTMADGDAVSIFGSSHIWVDHNS 953
Query: 189 MRNCRDGLIDAIMGSTAITISNSH 212
+ +C DGL+DA+MGSTAITISN+H
Sbjct: 954 LSHCADGLVDAVMGSTAITISNNH 1025
Score = 178 bits (451), Expect(2) = e-120
Identities = 83/161 (51%), Positives = 110/161 (67%)
Frame = +1
Query: 213 FTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMY 272
FT HNEV+L G SD+Y D++MQ+T+ +N FG+ LIQRMPRCR G+ HV+NN Y WEMY
Sbjct: 1027 FTHHNEVILLGHSDSYTRDKQMQVTIAYNHFGEGLIQRMPRCRHGYFHVVNNDYTHWEMY 1206
Query: 273 AIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQS 332
AIGG+ PTI S+GN++ AP N AKE+TKR E +WK W WRS DLYLNGA+F S
Sbjct: 1207 AIGGSADPTINSQGNRYNAPVNPFAKEVTKRVETAETQWKGWNWRSEGDLYLNGAYFTAS 1386
Query: 333 GAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
GA + ++ + AK + VG +T + +L C+ G+ C
Sbjct: 1387 GAG-ASASYARASSLGAKSSAMVGTMTSNAGALGCKRGRQC 1506
>TC85871 similar to GP|7547009|gb|AAF63756.1| pectate lyase {Vitis
vinifera}, partial (84%)
Length = 1481
Score = 350 bits (899), Expect(2) = e-114
Identities = 164/284 (57%), Positives = 208/284 (72%)
Frame = +1
Query: 90 PLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKIYDIM 149
PLWIIFAR M I+L +ELIM KTIDGRGA V IA G ITIQ++ N+IIHGI I+D
Sbjct: 253 PLWIIFARDMVIKLKEELIMNSFKTIDGRGASVHIAGGPCITIQYVTNIIIHGIHIHDCK 432
Query: 150 VGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGSTAITIS 209
G +VRDS H+G RTISDGDG+SIFG SH+W+DH S+ NC DGLIDAI GSTAITIS
Sbjct: 433 QGGNAMVRDSPRHFGWRTISDGDGVSIFGGSHVWVDHCSLSNCEDGLIDAIYGSTAITIS 612
Query: 210 NSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRW 269
N++ T H++VML G SD+Y D+ MQ+T+ FN FG++L+QRMPRCR G+ HV+NN Y W
Sbjct: 613 NNYMTHHDKVMLLGHSDSYTHDKNMQVTIAFNHFGERLVQRMPRCRLGYFHVVNNDYTHW 792
Query: 270 EMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLYLNGAFF 329
EMYAIGG+ +PTI S+GN+F+APNN +KE+TK E+EWK W WRS DL +NGAFF
Sbjct: 793 EMYAIGGSANPTINSQGNRFVAPNNRFSKEVTKYEDAAESEWKHWNWRSEGDLMVNGAFF 972
Query: 330 RQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
+SG + ++ + A+P S VG +T + +L C+ G PC
Sbjct: 973 TKSGGG-ASSSYARASSLSARPSSIVGSITIGAGTLNCKKGSPC 1101
Score = 80.1 bits (196), Expect(2) = e-114
Identities = 37/64 (57%), Positives = 43/64 (66%)
Frame = +3
Query: 3 IGGINGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPI 62
I + RRNL G SC + NPID CWRCD NW NR++LADC GFG+ GGK+G I
Sbjct: 72 INASSARRNL---GYLSCGSGNPIDDCWRCDSNWEKNRQRLADCAIGFGKNAIGGKNGKI 242
Query: 63 YVVT 66
YVVT
Sbjct: 243 YVVT 254
>TC89125 similar to PIR|H86253|H86253 hypothetical protein [imported] -
Arabidopsis thaliana, partial (70%)
Length = 1334
Score = 373 bits (957), Expect = e-104
Identities = 188/350 (53%), Positives = 234/350 (66%)
Frame = +3
Query: 24 NPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPGTLRH 83
N ID CWR PNWA NRK LADC GFG+ + GGK G IY+VTD SD D NP+PGTLR+
Sbjct: 120 NKIDSCWRAKPNWALNRKALADCAIGFGKDSIGGKYGAIYIVTDSSD-DPANPKPGTLRY 296
Query: 84 AVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGI 143
+ PLWIIF R+M + L ELIM KTIDGRG V I NG ITIQ + +VIIHGI
Sbjct: 297 GAIQTKPLWIIFERNMVLTLKNELIMNSYKTIDGRGVKVEIGNGPCITIQGVSHVIIHGI 476
Query: 144 KIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGS 203
I+D GLVR + DH G R +DGD ISIF SS+IWIDH + DGLID I S
Sbjct: 477 SIHDCKPSKAGLVRSTPDHVGRRRGADGDAISIFASSNIWIDHCFLARSTDGLIDIIHAS 656
Query: 204 TAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLN 263
TAITISN++FT H++VML G +D Y D+ M++T+VFNRFG LI+RMPR RFG+ HV+N
Sbjct: 657 TAITISNNYFTQHDKVMLLGHNDEYTADKIMKVTIVFNRFGSGLIERMPRVRFGYAHVVN 836
Query: 264 NFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLY 323
N Y++W+MYAIGG+ +PTI+SEGN + APN+ K+ITKR + WK+W+WRS D +
Sbjct: 837 NKYDQWQMYAIGGSANPTILSEGNFYNAPNDHTKKQITKRE--SKGNWKSWKWRSSKDYF 1010
Query: 324 LNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
NGA+F SG ++ A PG V +T + L C VG+ C
Sbjct: 1011SNGAYFIPSGYGSCAPNYTPAQSFVAVPGYMVPAITLNAGPLSCFVGRSC 1160
>TC90147 similar to GP|21537059|gb|AAM61400.1 pectate lyase-like protein
{Arabidopsis thaliana}, partial (55%)
Length = 1093
Score = 290 bits (743), Expect(2) = 5e-93
Identities = 137/219 (62%), Positives = 166/219 (75%)
Frame = +3
Query: 63 YVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADV 122
Y+VTD SD+D VNP+PGTLR+AV +N PLWI+F +M I+L+QELI KTIDGRGADV
Sbjct: 435 YIVTDSSDNDAVNPKPGTLRYAVIQNEPLWIVFPSNMMIKLSQELIFNSFKTIDGRGADV 614
Query: 123 TIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHI 182
I G IT+Q+I NVIIH I ++ VR S +HYG RT SDGDGISIFGS I
Sbjct: 615 HIVGGGCITLQYISNVIIHNIHVHHCHPSGNTNVRSSPEHYGYRTESDGDGISIFGSKDI 794
Query: 183 WIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNR 242
WIDH ++ C+DGLID +MGSTAITISN+HF+ HNEVML G SD Y D MQ+T+ FN
Sbjct: 795 WIDHCTLSRCKDGLIDVVMGSTAITISNNHFSHHNEVMLLGHSDHYKPDSAMQVTIAFNH 974
Query: 243 FGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPT 281
FG++L+QRMPRCR G+IHV+NN + RWEMYAIGG+ PT
Sbjct: 975 FGQQLVQRMPRCRLGYIHVVNNDFTRWEMYAIGGSGGPT 1091
Score = 68.9 bits (167), Expect(2) = 5e-93
Identities = 28/54 (51%), Positives = 38/54 (69%), Gaps = 4/54 (7%)
Frame = +1
Query: 9 RRNLVGLGN----SSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGK 58
RR ++ + N +SC+ NPID CW+CD +WANNR++LADC GFG+ GGK
Sbjct: 259 RREMLSISNKEFSTSCITGNPIDDCWKCDTDWANNRQRLADCAIGFGQNAKGGK 420
>TC87959 similar to SP|P24396|9612_LYCES Style development-specific protein
9612 precursor. [Tomato] {Lycopersicon esculentum},
partial (68%)
Length = 1340
Score = 323 bits (827), Expect = 9e-89
Identities = 154/279 (55%), Positives = 199/279 (71%)
Frame = +1
Query: 95 FARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKIYDIMVGSGG 154
F R M I+L QEL++ KTIDGRGA V IANG ITI ++ NVIIHGI ++D +
Sbjct: 1 FKRDMVIQLKQELLVNSYKTIDGRGASVHIANGGCITIHYVNNVIIHGIHVHDCVPTGNT 180
Query: 155 LVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGSTAITISNSHFT 214
+RDS +H G T+SDGDGIS+F S HIWIDH S+ NCRDGLID I GS AITISN++ T
Sbjct: 181 NIRDSPEHSGFWTVSDGDGISVFNSQHIWIDHCSLSNCRDGLIDVIHGSNAITISNNYMT 360
Query: 215 DHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAI 274
H++VML G SD+Y D+ MQ+T+ FN FG+ L+QRMPRCR G+ HV+NN Y WEMYAI
Sbjct: 361 HHDKVMLLGHSDSYTQDKDMQVTIAFNHFGEGLVQRMPRCRHGYFHVVNNDYTHWEMYAI 540
Query: 275 GGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGA 334
GG+ +PTI S+GN+F+AP+N +KE+TK E+E+ +W WRS DL+LNGAFFRQ+GA
Sbjct: 541 GGSANPTINSQGNRFLAPDNRFSKEVTKHEDASESEYNSWNWRSEGDLFLNGAFFRQTGA 720
Query: 335 ELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
E + ++ + A+P S VG +T S L C+ G C
Sbjct: 721 E-SSSIYARASSLSARPASLVGSITTTSGVLTCKKGNRC 834
>TC85870 similar to GP|18087517|gb|AAL58893.1 At1g67750/F12A21_12
{Arabidopsis thaliana}, partial (73%)
Length = 1521
Score = 322 bits (826), Expect = 1e-88
Identities = 155/251 (61%), Positives = 186/251 (73%)
Frame = +2
Query: 3 IGGINGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPI 62
I + RRNL G SC + NPID CWRCD NW NR++LADC GFG+ GGK+G I
Sbjct: 167 INASSARRNL---GYLSCGSGNPIDDCWRCDSNWEKNRQRLADCAIGFGKNAIGGKNGKI 337
Query: 63 YVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADV 122
YVVTD SD + V P+PGTLRHAV + PLWIIFAR M I+L +ELIM KTIDGRGA V
Sbjct: 338 YVVTDASDDNPVTPKPGTLRHAVIQVEPLWIIFARDMVIKLKEELIMNSFKTIDGRGASV 517
Query: 123 TIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHI 182
IA G ITIQ++ N+IIHGI I+D G +VRDS H+G RTISDGDG+SIFG SH+
Sbjct: 518 HIAGGPCITIQYVTNIIIHGIHIHDCKQGGNAMVRDSPRHFGWRTISDGDGVSIFGGSHV 697
Query: 183 WIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNR 242
W+DH S+ NC DGLIDAI GSTAITISN++ T H++VML G SD+Y D+ MQ+T+ FN
Sbjct: 698 WVDHCSLSNCEDGLIDAIYGSTAITISNNYMTHHDKVMLLGHSDSYTHDKNMQVTIAFNH 877
Query: 243 FGKKLIQRMPR 253
FG+ L+QRMPR
Sbjct: 878 FGEGLVQRMPR 910
Score = 57.0 bits (136), Expect = 1e-08
Identities = 26/52 (50%), Positives = 34/52 (65%)
Frame = +3
Query: 235 QITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEG 286
Q+ + +R K L RCR G+ HV+NN Y WEMYAIGG+ +PTI S+G
Sbjct: 1365 QVRALVSRI*KNL*IFFYRCRLGYFHVVNNDYTHWEMYAIGGSANPTINSQG 1520
>BQ147652 homologue to PIR|T09524|T09 probable pectate lyase (EC 4.2.2.2) -
alfalfa, partial (46%)
Length = 716
Score = 275 bits (702), Expect = 3e-74
Identities = 128/209 (61%), Positives = 155/209 (73%)
Frame = +1
Query: 165 LRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGA 224
+RT SDGD IS+FGSS+IWIDH+S+ NC DGL+D I GSTA+TISN H T HN+VMLFGA
Sbjct: 1 VRTRSDGDAISVFGSSNIWIDHISLSNCEDGLVDVIQGSTAVTISNCHMTKHNDVMLFGA 180
Query: 225 SDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIIS 284
SDTY D+ MQ+TV FN FG+ LIQRMPRCR+GF HVLNN Y W MYAIGG+ PTI+S
Sbjct: 181 SDTYQDDKIMQVTVAFNHFGQGLIQRMPRCRWGFFHVLNNDYTHWIMYAIGGSSAPTILS 360
Query: 285 EGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNK 344
+GN+FIAP+N AK IT R PEA W WQWRS D ++NGA F QSG + + PF
Sbjct: 361 QGNRFIAPHNNAAKTITHRDYAPEAVWSKWQWRSEGDHFMNGANFIQSGPPIKSLPFKKG 540
Query: 345 DMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
++K + GS LTR+S +L C VG+PC
Sbjct: 541 FLMKPRHGSXANXLTRFSGALNCVVGRPC 627
>BI272128 similar to PIR|T09524|T09 probable pectate lyase (EC 4.2.2.2) -
alfalfa, partial (47%)
Length = 682
Score = 250 bits (638), Expect(2) = 4e-71
Identities = 122/191 (63%), Positives = 148/191 (76%), Gaps = 7/191 (3%)
Frame = +3
Query: 87 RNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKIY 146
+ GPLWI FARSM IRLNQEL+++ +KTIDGRGA+V I +GAGIT+QF+ NVIIHG+ I
Sbjct: 69 QKGPLWITFARSMVIRLNQELMVSSDKTIDGRGANVQIRDGAGITMQFVNNVIIHGLHIQ 248
Query: 147 DIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGSTAI 206
+I GGL+RDS DH G RT SDGD ISI+GSS+IWIDH+S+ C DGL+D I GST I
Sbjct: 249 NIKAKPGGLIRDSFDHTGQRTRSDGDAISIYGSSNIWIDHLSLSECEDGLVDIIQGSTGI 428
Query: 207 TISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGF-------I 259
TISN H T HN+VMLFGASD+Y GD+ MQITV FN FG+ LIQRMPRCR+GF +
Sbjct: 429 TISNCHMTKHNDVMLFGASDSYTGDKLMQITVAFNHFGQGLIQRMPRCRYGFCSCFEQ*L 608
Query: 260 HVLNNFYNRWE 270
+ L+N N W+
Sbjct: 609 YSLDNVCNWWK 641
Score = 39.3 bits (90), Expect = 0.002
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = +1
Query: 257 GFIHVLNNFYNRWEMYAIGGTM 278
GF+HVLNN Y W MYAIGG++
Sbjct: 580 GFVHVLNNDYTHWIMYAIGGSL 645
Score = 36.2 bits (82), Expect(2) = 4e-71
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +2
Query: 65 VTDPSDSDLVNPRPGTLRHAVTRNGPL 91
VTD SD+D+VNP+PGTLR G +
Sbjct: 2 VTDESDNDMVNPKPGTLRFGACPKGTI 82
>TC85009 similar to GP|14531294|gb|AAK66160.1 pectate lyase B {Fragaria x
ananassa}, partial (55%)
Length = 914
Score = 238 bits (608), Expect = 2e-63
Identities = 119/188 (63%), Positives = 134/188 (70%)
Frame = +3
Query: 19 SCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRP 78
SC NP+D CWRCD W RK+LADC GFGR GG+DG YVV +P D D VNPRP
Sbjct: 351 SCGTGNPMDDCWRCDKLWYRRRKRLADCAIGFGRNAIGGRDGRYYVVNNPRDDDPVNPRP 530
Query: 79 GTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENV 138
GTLRHAV ++ PLWI+F R M I L QELIM KTIDGRGA+V IA GA ITIQFI NV
Sbjct: 531 GTLRHAVIQDRPLWIVFKRDMVITLKQELIMNSFKTIDGRGANVHIAFGACITIQFITNV 710
Query: 139 IIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLID 198
IIHG+ I+D +VR S H+G RT++DGDGISIFG SHIW S+ NC DGL D
Sbjct: 711 IIHGVHIHDCKPTGNAMVRSSPFHFGWRTMADGDGISIFGFSHIWD*PHSLSNCADGLCD 890
Query: 199 AIMGSTAI 206
AIMGSTAI
Sbjct: 891 AIMGSTAI 914
>TC90880 similar to GP|15027941|gb|AAK76501.1 unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 1051
Score = 159 bits (402), Expect = 2e-39
Identities = 74/75 (98%), Positives = 75/75 (99%)
Frame = -2
Query: 299 EITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRL 358
+ITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRL
Sbjct: 1014 KITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRL 835
Query: 359 TRYSRSLRCRVGKPC 373
TRYSRSLRCRVGKPC
Sbjct: 834 TRYSRSLRCRVGKPC 790
>AW688239 similar to GP|21537059|gb pectate lyase-like protein {Arabidopsis
thaliana}, partial (23%)
Length = 625
Score = 147 bits (372), Expect = 5e-36
Identities = 67/110 (60%), Positives = 78/110 (70%)
Frame = +1
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
L S+CM NPID CW+CDP W NNR++LADC GFG+ GGK G YVVTD SD D V
Sbjct: 268 LSGSACMTGNPIDDCWKCDPEWPNNRQRLADCAIGFGQYAKGGKGGEFYVVTDSSDDDAV 447
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTI 124
P+PGTLR+ V +N PLWI+F +M I+L QELI KTID RGADV I
Sbjct: 448 TPKPGTLRYGVIKNEPLWIVFPXNMMIKLKQELIFNSYKTIDXRGADVHI 597
>BG645082 similar to GP|20466073|gb At5g04300/At5g04300 {Arabidopsis
thaliana}, partial (41%)
Length = 782
Score = 147 bits (372), Expect = 5e-36
Identities = 65/140 (46%), Positives = 96/140 (68%)
Frame = +3
Query: 223 GASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTI 282
G +D Y D+ MQ+T+ FN FG+ L+QRMPRCR G+IHV+NN + +WEMYAIGG+ +PTI
Sbjct: 6 GHNDKYTPDRGMQVTIAFNHFGEGLVQRMPRCRLGYIHVVNNDFTQWEMYAIGGSANPTI 185
Query: 283 ISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFS 342
S+GN++ AP++ +AKE+TKR + EW W WR+ D+ +NGAFF SG + + ++
Sbjct: 186 NSQGNRYTAPSDANAKEVTKRVDTDDKEWSGWNWRTEGDIMVNGAFFVPSGVGM-SAQYA 362
Query: 343 NKDMIKAKPGSYVGRLTRYS 362
++ K + +LT YS
Sbjct: 363 EASSVQPKSVEQINQLTMYS 422
>BF004635 similar to PIR|H86253|H86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (30%)
Length = 538
Score = 135 bits (341), Expect = 2e-32
Identities = 66/122 (54%), Positives = 83/122 (67%)
Frame = +3
Query: 26 IDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPGTLRHAV 85
+ WR + NW +NR+ LADC GFG+ GGK G IY VTDPSD D +NP+ GTLR+ V
Sbjct: 174 LTHAWRSNSNWVSNRQSLADCAIGFGKDAIGGKYGEIYEVTDPSD-DPINPKKGTLRYGV 350
Query: 86 TRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKI 145
+ P+WIIF+ M I+L ELI+ KTIDGRGA V I NG ITIQ + +VI HGI +
Sbjct: 351 IQAEPVWIIFSNDMVIKLKNELIVNSYKTIDGRGAKVEIGNGPCITIQGVSHVIAHGISV 530
Query: 146 YD 147
+D
Sbjct: 531 HD 536
>TC90888 similar to GP|14289169|dbj|BAB59066. pectate lyase {Salix
gilgiana}, partial (31%)
Length = 481
Score = 128 bits (322), Expect = 3e-30
Identities = 62/103 (60%), Positives = 73/103 (70%)
Frame = +1
Query: 7 NGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVT 66
+ RRNL L SC + NPID CWRCD NW NR++LADC GFG+ GG+DG IYVVT
Sbjct: 181 DSRRNLAFL---SCGSGNPIDDCWRCDKNWEKNRQRLADCAIGFGKHAIGGRDGKIYVVT 351
Query: 67 DPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIM 109
DP D VNP+PGTLR+ V + PLWIIF R M I+L QEL+M
Sbjct: 352 DPGD-HAVNPKPGTLRYGVIQEEPLWIIFKRDMVIKLKQELMM 477
>BF519432 similar to GP|6682233|gb| putative pectate lyase {Arabidopsis
thaliana}, partial (45%)
Length = 531
Score = 103 bits (257), Expect = 1e-22
Identities = 70/184 (38%), Positives = 95/184 (51%), Gaps = 1/184 (0%)
Frame = +2
Query: 41 KKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMK 100
+ LA +GFGR GG G +Y VT L++ PG+LR A + PLWI+F S
Sbjct: 50 RSLAAQAEGFGRCAIGGLHGSLYHVTS-----LLDDGPGSLRDACRKKEPLWIVFEVSGT 214
Query: 101 IRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSE 160
I L+ L ++ KTIDGRG + + G G+ ++ E+VII ++I G G
Sbjct: 215 IHLSSYLSVSSYKTIDGRGQKIKLT-GKGLRLKECEHVIICNLEIEG---GRG------- 361
Query: 161 DHYGLRTISDGDGISIF-GSSHIWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEV 219
D D I I S HIWID ++ + DGLID GST ITIS HF H++
Sbjct: 362 --------PDVDAIQIKPNSKHIWIDRCTLSDFEDGLIDITRGSTDITISRCHFHQHDKT 517
Query: 220 MLFG 223
+L G
Sbjct: 518 ILIG 529
>BI273096 similar to GP|18087517|gb At1g67750/F12A21_12 {Arabidopsis
thaliana}, partial (25%)
Length = 359
Score = 100 bits (249), Expect = 9e-22
Identities = 44/73 (60%), Positives = 50/73 (68%)
Frame = +2
Query: 6 INGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVV 65
IN R LG SC NPID CWRCDPNW NNR++LADC GFG+ GGK+G IY+V
Sbjct: 140 INVSRXRRNLGYLSCGTGNPIDDCWRCDPNWENNRQRLADCAIGFGKDAIGGKNGKIYIV 319
Query: 66 TDPSDSDLVNPRP 78
TD D D VNP+P
Sbjct: 320 TDSGDDDAVNPKP 358
>BF518692 similar to PIR|S51509|S515 pectase lyase - Aspergillus sp., partial
(7%)
Length = 361
Score = 31.6 bits (70), Expect = 0.52
Identities = 33/147 (22%), Positives = 52/147 (34%)
Frame = +1
Query: 40 RKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSM 99
R D + G+ TTGG G VT S G ++ + +G +
Sbjct: 16 RAAAIDQLVGYAAGTTGGGSGSGTTVTSCSALTTAAKAGGVIKISGNLSG------CGII 177
Query: 100 KIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDS 159
K+ N ++ G+ + G G I+ + NVI+ +K +D G
Sbjct: 178 KLVANTSVLGVGSNSG---------LTGGGFQIRKVSNVIVRNLKFHDAPDGK------- 309
Query: 160 EDHYGLRTISDGDGISIFGSSHIWIDH 186
D I I S+ +WIDH
Sbjct: 310 ------------DQIDIDASTKVWIDH 354
>TC77153 homologue to GP|21902507|gb|AAM78552.1 ribosomal protein small
subunit 28 kDa {Helianthus annuus}, complete
Length = 849
Score = 27.3 bits (59), Expect = 9.7
Identities = 13/34 (38%), Positives = 17/34 (49%)
Frame = +1
Query: 35 NWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
++ +KK G +KTTG DGP YV P
Sbjct: 682 SFLKKKKKKKKNRHGIRQKTTGWSDGPAYVTQCP 783
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,648,906
Number of Sequences: 36976
Number of extensions: 155523
Number of successful extensions: 770
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 761
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 764
length of query: 373
length of database: 9,014,727
effective HSP length: 98
effective length of query: 275
effective length of database: 5,391,079
effective search space: 1482546725
effective search space used: 1482546725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136286.13


