

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.11 - phase: 0
(620 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
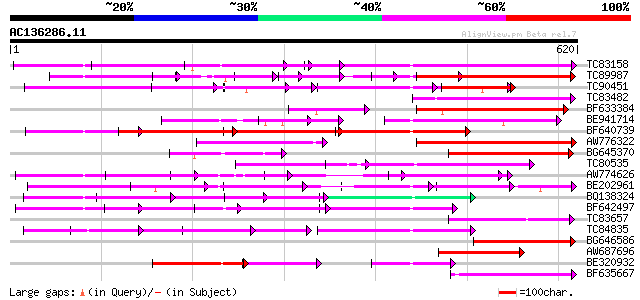
Score E
Sequences producing significant alignments: (bits) Value
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 182 3e-46
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 124 2e-40
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 111 5e-33
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 128 6e-30
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 127 1e-29
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 125 5e-29
BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis... 120 1e-27
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 115 4e-26
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 112 4e-25
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 102 4e-22
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 99 6e-21
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 99 6e-21
BQ138324 weakly similar to GP|12322902|gb| hypothetical protein;... 96 3e-20
BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T... 96 3e-20
TC83657 weakly similar to GP|11994382|dbj|BAB02341. gb|AAB82628.... 96 5e-20
TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein... 93 3e-19
BG646586 weakly similar to GP|10177683|dbj selenium-binding prot... 89 4e-18
AW687696 weakly similar to PIR|T49965|T499 hypothetical protein ... 81 1e-15
BE320932 weakly similar to PIR|T04548|T045 hypothetical protein ... 80 3e-15
BF635667 weakly similar to PIR|C85359|C853 hypothetical protein ... 79 5e-15
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 182 bits (462), Expect = 3e-46
Identities = 100/299 (33%), Positives = 168/299 (55%), Gaps = 2/299 (0%)
Frame = +2
Query: 323 AVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLS 382
A++LF+++ + ++ +W +I GF +K EA + F +MQ AGV P + +++S
Sbjct: 17 ALKLFDKLPVKNVV----SWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIIS 184
Query: 383 VCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDP 442
C + L +H ++ + + +L+D Y +CGC+ AR VFD + +
Sbjct: 185 ACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQR--NL 358
Query: 443 AFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRM 502
WN++I G+ NG + A F M E ++PN ++ S L+ACSH+G I+ GL+ F
Sbjct: 359 VSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFAD 538
Query: 503 IRK-YGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAEPPASV-FDSLLGACRCYLDSN 560
I++ + P+ EH+GC+VDL RAG+L EA D+++++ P V SLL ACR D
Sbjct: 539 IKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVE 718
Query: 561 LGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
L E++ +++ P + V+ SNIYAA+G+W ++R + ++GL KN S IE+
Sbjct: 719 LAEKVMKYQVELYPGGDSNYVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFSSIEI 895
Score = 122 bits (307), Expect = 3e-28
Identities = 61/175 (34%), Positives = 105/175 (59%)
Frame = +2
Query: 192 KVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATL 251
K+F+ L VKNVV++ + G ++ + + F++M + P+ VT+++++SACA L
Sbjct: 23 KLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQL-AGVVPDFVTVIAIISACANL 199
Query: 252 SNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMI 311
+ LG VH L MK E D+V V+ SL+DMY++CGC A VF +RNL++WNS+I
Sbjct: 200 GALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSWNSII 379
Query: 312 AGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQ 366
G +N +++A+ F M EG+ P+ ++ S ++ + G+ E K F+ ++
Sbjct: 380 VGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIK 544
Score = 84.7 bits (208), Expect = 9e-17
Identities = 61/246 (24%), Positives = 118/246 (47%), Gaps = 3/246 (1%)
Frame = +2
Query: 90 ALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA-RD 148
AL+LFD++P + ++ V+ G + +A+ FR++ + P+ VT+++++SA +
Sbjct: 17 ALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISACAN 196
Query: 149 VKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAF 208
+ VH L K +V V SL+ Y++CG + + +VF+ + +N+V++N+
Sbjct: 197 LGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSWNSI 376
Query: 209 MSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHG-LSMKL 267
+ G NG F+ M E PN V+ S ++AC+ I G ++ +
Sbjct: 377 IVGFAVNGLADKALSFFRSMKKEGLE-PNGVSYTSALTACSHAGLIDEGLKIFADIKRDH 553
Query: 268 EACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKR-NLITWNSMIAGMMMNSESERAVEL 326
+ LVD+YS+ G A+DV + N + S++A + E A ++
Sbjct: 554 RNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVELAEKV 733
Query: 327 FERMVD 332
+ V+
Sbjct: 734 MKYQVE 751
Score = 73.9 bits (180), Expect = 2e-13
Identities = 71/306 (23%), Positives = 127/306 (41%), Gaps = 6/306 (1%)
Frame = +2
Query: 5 ITVTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKT 64
+ + V Y+EAL + + + P+ T ++ AC+NL + +H + K
Sbjct: 68 VVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISACANLGALGLGLWVHRLVMKK 247
Query: 65 GFHSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWL 124
F + +LI Y A A ++FD M Q + ++N+++ G + NG +A+
Sbjct: 248 EFRDNVKVLNSLIDMY-ARCGCIELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKALSF 424
Query: 125 FRQIGFWNIRPNSVTIVSLLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKC 184
FR + + PN GV Y TS +TA S
Sbjct: 425 FRSMKKEGLEPN-----------------------------GVSY-----TSALTACSHA 502
Query: 185 GVLVSSNKVFENLR-----VKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKV 239
G++ K+F +++ + Y + + G + +DV K M M PN+V
Sbjct: 503 GLIDEGLKIFADIKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPM----MPNEV 670
Query: 240 TLVSVVSACATLSNIRLGKQVHGLSMKL-EACDHVMVVTSLVDMYSKCGCWGSAFDVFSR 298
L S+++AC T ++ L ++V ++L D V+ S ++Y+ G W A V
Sbjct: 671 VLGSLLAACRTQGDVELAEKVMKYQVELYPGGDSNYVLFS--NIYAAVGKWDGASKVRRE 844
Query: 299 SEKRNL 304
++R L
Sbjct: 845 MKERGL 862
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 124 bits (312), Expect(2) = 2e-40
Identities = 62/176 (35%), Positives = 109/176 (61%), Gaps = 2/176 (1%)
Frame = +2
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIR 504
+ MI G +G + A +VF EM++E + P+ +V V SACSH+G +E GL+ F+ ++
Sbjct: 617 YTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHAGLVEEGLQCFKSMQ 796
Query: 505 -KYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLG 562
++ ++P +H+GC+VDLLGR G L EA +L++ ++ +P ++ SLL AC+ + + +G
Sbjct: 797 FEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWRSLLSACKVHHNLEIG 976
Query: 563 EEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIE 618
+ A L + N +VL+N+YA +W +V +IR + ++ L + G S+IE
Sbjct: 977 KIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRTKLAERNLVQTPGFSLIE 1144
Score = 76.6 bits (187), Expect(2) = 3e-22
Identities = 50/144 (34%), Positives = 80/144 (54%), Gaps = 7/144 (4%)
Frame = +1
Query: 157 QVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNG 216
QVH K+G+E DV V SL+ Y KCG + ++ VF + K+V +++A + G
Sbjct: 166 QVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVASWSAII------G 327
Query: 217 FHRVVFDVFKDMTMNLEE-------KPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEA 269
H V +++ + M L + + + TLV+V+SAC L + LGK +HG+ ++ +
Sbjct: 328 AHACV-EMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGILLRNIS 504
Query: 270 CDHVMVVTSLVDMYSKCGCWGSAF 293
+V+V TSL+DMY K GC F
Sbjct: 505 ELNVVVKTSLIDMYVKSGCLEKRF 576
Score = 60.1 bits (144), Expect(2) = 2e-40
Identities = 48/180 (26%), Positives = 81/180 (44%), Gaps = 1/180 (0%)
Frame = +1
Query: 247 ACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLIT 306
AC+ L + G QVHG K+ V+V SL++MY KCG +A DVF+ +++++ +
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 307 WNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVC-VEAFKYFSKM 365
W+++I + VE++ + L+ + +G C VE
Sbjct: 310 WSAIIG-------AHACVEMWNECL------------MLLGKMSSEGRCRVEE------- 411
Query: 366 QCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCV 425
L ++LS C K IHG LR + + + T+L+D Y+K GC+
Sbjct: 412 ---------STLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGCL 564
Score = 50.1 bits (118), Expect = 3e-06
Identities = 30/102 (29%), Positives = 53/102 (51%), Gaps = 1/102 (0%)
Frame = +1
Query: 396 IHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTN 455
+HG+ ++ ++ D + +L++ Y KCG + A VF+ D K A W+A+IG +
Sbjct: 169 VHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEK--SVASWSAIIGAHACV 342
Query: 456 GDYESAFEVFYEMLDE-MVQPNSATFVSVLSACSHSGQIERG 496
+ + +M E + +T V+VLSAC+H G + G
Sbjct: 343 EMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLG 468
Score = 47.4 bits (111), Expect(2) = 9e-11
Identities = 39/144 (27%), Positives = 67/144 (46%), Gaps = 4/144 (2%)
Frame = +2
Query: 182 SKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEE--KPNKV 239
S+ VL +VF+N+ KN +Y +SGL +G + VF +M +EE P+ V
Sbjct: 548 SRVDVLRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEM---IEEGLAPDDV 718
Query: 240 TLVSVVSACATLSNIRLGKQ-VHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVF-S 297
V V SAC+ + G Q + + + V +VD+ + G A+++ S
Sbjct: 719 VYVGVFSACSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKS 898
Query: 298 RSEKRNLITWNSMIAGMMMNSESE 321
S K N + W S+++ ++ E
Sbjct: 899 MSIKPNDVIWRSLLSACKVHHNLE 970
Score = 47.0 bits (110), Expect(2) = 3e-22
Identities = 19/72 (26%), Positives = 39/72 (53%)
Frame = +2
Query: 295 VFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGV 354
VF ++N ++ MI+G+ ++ + A+++F M++EG+ PD + + S + G+
Sbjct: 581 VFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHAGL 760
Query: 355 CVEAFKYFSKMQ 366
E + F MQ
Sbjct: 761 VEEGLQCFKSMQ 796
Score = 41.6 bits (96), Expect = 0.001
Identities = 48/218 (22%), Positives = 91/218 (41%), Gaps = 3/218 (1%)
Frame = +2
Query: 91 LELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARDVK 150
L +F M + ++ ++SGL+ +G +A+ +F ++ + P+ V V + SA
Sbjct: 575 LRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSA---- 742
Query: 151 NQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMS 210
C L E G+ + FE+ V Y +
Sbjct: 743 ---------CSHAGLVEE----------------GLQCFKSMQFEHKIEPTVQHYGCMVD 847
Query: 211 GLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGK-QVHGLSM--KL 267
L + G + +++ K M++ KPN V S++SAC N+ +GK L M +
Sbjct: 848 LLGRFGMLKEAYELIKSMSI----KPNDVIWRSLLSACKVHHNLEIGKIAAENLFMLNQN 1015
Query: 268 EACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLI 305
+ D+++ L +MY+K W + ++ +RNL+
Sbjct: 1016NSGDYLV----LANMYAKAQKWDDVAKIRTKLAERNLV 1117
Score = 37.4 bits (85), Expect(2) = 9e-11
Identities = 34/146 (23%), Positives = 65/146 (44%), Gaps = 2/146 (1%)
Frame = +1
Query: 44 ACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEMPQPTIT 103
ACS L + +H H+FK G +LI Y A ++F+ M + ++
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMY-GKCGEIKNACDVFNGMDEKSVA 306
Query: 104 AFNAVLSGLSRNGPRGQAVWLFRQIGF-WNIRPNSVTIVSLLSA-RDVKNQSHVQQVHCL 161
+++A++ + + + L ++ R T+V++LSA + + + +H +
Sbjct: 307 SWSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGI 486
Query: 162 ACKLGVEYDVYVSTSLVTAYSKCGVL 187
+ E +V V TSL+ Y K G L
Sbjct: 487 LLRNISELNVVVKTSLIDMYVKSGCL 564
Score = 33.9 bits (76), Expect = 0.19
Identities = 38/157 (24%), Positives = 61/157 (38%), Gaps = 1/157 (0%)
Frame = +2
Query: 341 TWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYA 400
++ +ISG A G EA K FS+M G+AP + + S C +
Sbjct: 614 SYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHA------------- 754
Query: 401 LRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPA-FWNAMIGGYGTNGDYE 459
LV+ ++C F QF+ K + + M+ G G +
Sbjct: 755 -------------GLVEEGLQC-------FKSMQFEHKIEPTVQHYGCMVDLLGRFGMLK 874
Query: 460 SAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERG 496
A+E+ M ++PN + S+LSAC +E G
Sbjct: 875 EAYELIKSM---SIKPNDVIWRSLLSACKVHHNLEIG 976
Score = 32.7 bits (73), Expect = 0.42
Identities = 33/154 (21%), Positives = 60/154 (38%)
Frame = +2
Query: 5 ITVTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKT 64
+ ++ L +G KEAL ++S + P+ + + ACS HA L +
Sbjct: 623 VMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACS-----------HAGLVEE 769
Query: 65 GFHSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWL 124
G + F+ +PT+ + ++ L R G +A L
Sbjct: 770 GLQC--------------------FKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYEL 889
Query: 125 FRQIGFWNIRPNSVTIVSLLSARDVKNQSHVQQV 158
+ + +I+PN V SLLSA V + + ++
Sbjct: 890 IKSM---SIKPNDVIWRSLLSACKVHHNLEIGKI 982
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 111 bits (277), Expect = 9e-25
Identities = 62/182 (34%), Positives = 101/182 (55%), Gaps = 2/182 (1%)
Frame = +2
Query: 156 QQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQN 215
QQVH KLG++ + Y + + Y K G S++KVF+ + ++NA +SGL Q
Sbjct: 545 QQVHSYGIKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQG 724
Query: 216 GFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMV 275
G VF DM + E P+ +T+VSV+SAC ++ ++ L Q+H + + + ++
Sbjct: 725 GLAMDAIVVFVDMKRHGFE-PDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVI 901
Query: 276 V--TSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDE 333
+ SL+DMY KCG A++VF+ E RN+ +W SMI G M+ ++ A+ F M
Sbjct: 902 LMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCMKGV 1081
Query: 334 GI 335
G+
Sbjct: 1082GV 1087
Score = 83.2 bits (204), Expect(2) = 2e-18
Identities = 57/215 (26%), Positives = 105/215 (48%), Gaps = 3/215 (1%)
Frame = +2
Query: 17 KEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTAL 76
+ AL +Y + + P+ +T PI+LKA S + Q +H++ K G S+ + +
Sbjct: 431 QNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGF 610
Query: 77 IASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPN 136
I Y F A ++FDE +P + ++NA++SGLS+ G A+ +F + P+
Sbjct: 611 INLYC-KAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPD 787
Query: 137 SVTIVSLLSA-RDVKNQSHVQQVH--CLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKV 193
+T+VS++SA + + Q+H K + +S SL+ Y KCG + + +V
Sbjct: 788 GITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEV 967
Query: 194 FENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDM 228
F + +NV ++ + + G +G + F M
Sbjct: 968 FATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 80.1 bits (196), Expect(2) = 5e-33
Identities = 37/82 (45%), Positives = 54/82 (65%), Gaps = 1/82 (1%)
Frame = +1
Query: 473 VQPNSATFVSVLSACSHSGQIERGLRFFRMIRK-YGLDPKPEHFGCVVDLLGRAGQLGEA 531
V+PN TF+ VLSAC H G ++ G +F M++ YG+ P+ +H+GC+VDLLGRAG +A
Sbjct: 1087 VKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDDA 1266
Query: 532 RDLVQELAEPPASVFDSLLGAC 553
R +V+E+ P SV + C
Sbjct: 1267 RRMVEEMPMKPNSVVWGVFDGC 1332
Score = 79.7 bits (195), Expect(2) = 5e-33
Identities = 62/235 (26%), Positives = 102/235 (43%), Gaps = 2/235 (0%)
Frame = +2
Query: 236 PNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDV 295
P++ TL V+ A + I+LG+QVH +KL + + +++Y K G + SA V
Sbjct: 479 PDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFINLYCKAGDFDSAHKV 658
Query: 296 FSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVC 355
F DE P +WN+LISG +Q G+
Sbjct: 659 F-----------------------------------DENHEPKLGSWNALISGLSQGGLA 733
Query: 356 VEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDD--FLAT 413
++A F M+ G P + S++S CG L A +H Y + ++ ++
Sbjct: 734 MDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSN 913
Query: 414 ALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEM 468
+L+D Y KCG + A VF + + + + W +MI GY +G + A F+ M
Sbjct: 914 SLIDMYGKCGRMDLAYEVFATMEDR--NVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 79.0 bits (193), Expect = 5e-15
Identities = 53/222 (23%), Positives = 98/222 (43%), Gaps = 8/222 (3%)
Frame = +2
Query: 337 PDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAI 396
P S WN++I + + A + + M AGV P L +L S ++ + +
Sbjct: 374 PASFNWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQV 553
Query: 397 HGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAF--WNAMIGGYGT 454
H Y +++ + +++ + ++ Y K G A VFD+ +P WNA+I G
Sbjct: 554 HSYGIKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDE----NHEPKLGSWNALISGLSQ 721
Query: 455 NGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEH 514
G A VF +M +P+ T VSV+SAC G + L+ + KY K
Sbjct: 722 GGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQ----LHKYVFQAKTNE 889
Query: 515 F------GCVVDLLGRAGQLGEARDLVQELAEPPASVFDSLL 550
+ ++D+ G+ G++ A ++ + + S + S++
Sbjct: 890 WTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMI 1015
Score = 27.7 bits (60), Expect(2) = 2e-18
Identities = 20/81 (24%), Positives = 36/81 (43%), Gaps = 8/81 (9%)
Frame = +1
Query: 235 KPNKVTLVSVVSACATLSNIRLG-------KQVHGLSMKLEACDHVMVVTSLVDMYSKCG 287
KPN VT + V+SAC ++ G K ++G++ +L+ +VD+ + G
Sbjct: 1090 KPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQH------YGCMVDLLGRAG 1251
Query: 288 CWGSAFDVFSRSE-KRNLITW 307
+ A + K N + W
Sbjct: 1252 LFDDARRMVEEMPMKPNSVVW 1314
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 128 bits (322), Expect = 6e-30
Identities = 69/180 (38%), Positives = 106/180 (58%), Gaps = 2/180 (1%)
Frame = +2
Query: 441 DPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFF 500
D WNA+IGGY ++G A E F + + + P+ TFV+VLSAC H+G +E G + F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEF-DRMKVVGTPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 501 R-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLD 558
M+ KYG+ + EH+ C+VDL GRAG+ EA +L++ + EP ++ +LL AC + +
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 559 SNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIE 618
LGE A ++ +E +P VLS I G WS V +R + ++G+ K + IS +E
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISWVE 538
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/63 (33%), Positives = 37/63 (58%)
Frame = +2
Query: 303 NLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYF 362
+L++WN++I G + + RA+E F+RM G PD T+ +++S G+ E K+F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVG-TPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 363 SKM 365
+ M
Sbjct: 179 TDM 187
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/122 (20%), Positives = 48/122 (38%)
Frame = +2
Query: 338 DSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIH 397
D +WN++I G+A G+ A + F +M+ G + + L + V K
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGTPDEVTFVNVLSACVHAGLVEEGEKHFT 181
Query: 398 GYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGD 457
+ + + + +VD Y + G A + +P D W A++ G + +
Sbjct: 182 DMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEP-DVVLWGALLAACGLHSN 358
Query: 458 YE 459
E
Sbjct: 359 LE 364
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 127 bits (319), Expect = 1e-29
Identities = 63/175 (36%), Positives = 108/175 (61%), Gaps = 8/175 (4%)
Frame = +1
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDE-----MVQPNSATFVSVLSACSHSGQIERGLR- 498
WN +I YG +G E A ++F M++E ++PN T++++ ++ SHSG ++ GL
Sbjct: 7 WNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGLNL 186
Query: 499 FFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAEPPASV--FDSLLGACRCY 556
F+ M K+G++P +H+ C+VDLLGR+GQ+ EA +L++ + V + SLLGAC+ +
Sbjct: 187 FYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACKIH 366
Query: 557 LDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKN 611
+ +GE A L ++P + V+LSNIY++ G W + +R + +KG+ KN
Sbjct: 367 QNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRKN 531
Score = 45.4 bits (106), Expect = 6e-05
Identities = 28/95 (29%), Positives = 46/95 (47%), Gaps = 6/95 (6%)
Frame = +1
Query: 305 ITWNSMIAGMMMNSESERAVELFERMVDEG-----ILPDSATWNSLISGFAQKGVCVEAF 359
ITWN +I M+ + E A++LF RMV+EG I P+ T+ ++ + + G+ E
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 360 KYFSKMQCA-GVAPCLKILTSLLSVCGDSCVLRSA 393
F M+ G+ P L+ + G S + A
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEA 285
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 125 bits (314), Expect = 5e-29
Identities = 69/200 (34%), Positives = 117/200 (58%), Gaps = 7/200 (3%)
Frame = +2
Query: 411 LATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLD 470
L+T+L+D Y KCG + A+ +FD +++ D WNAMI G +GD + A ++FY+M
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMR--DVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 471 EMVQPNSATFVSVLSACSHSGQIERGLRFF-RMIRKYGLDPKPEHFGCVVDLLGRAGQLG 529
V+P+ TF++V +ACS+SG GL +M Y + PK EH+GC+VDLL RAG
Sbjct: 194 VGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAGLFE 373
Query: 530 EARDLVQEL-----AEPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPK-NPAPLVVL 583
EA +++++ + + L AC + ++ L E A K++ ++ + V+L
Sbjct: 374 EAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGVYVLL 553
Query: 584 SNIYAALGRWSEVERIRGLI 603
SN+YAA G+ ++ R++ ++
Sbjct: 554 SNLYAASGKHNDARRVKDMM 613
Score = 65.1 bits (157), Expect = 8e-11
Identities = 32/93 (34%), Positives = 53/93 (56%)
Frame = +2
Query: 273 VMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVD 332
V + TSL+DMY+KCG A +F R+++ WN+MI+GM M+ + + A++LF M
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 333 EGILPDSATWNSLISGFAQKGVCVEAFKYFSKM 365
G+ PD T+ ++ + + G+ E KM
Sbjct: 194 VGVKPDDITFIAVFTACSYSGMAYEGLMLLDKM 292
Score = 63.5 bits (153), Expect = 2e-10
Identities = 44/176 (25%), Positives = 87/176 (49%), Gaps = 11/176 (6%)
Frame = +2
Query: 167 VEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFK 226
V V +STSL+ Y+KCG L + ++F+++ +++VV +NA +SG+ +G + +F
Sbjct: 2 VPLSVRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFY 181
Query: 227 DMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTS------LV 280
DM + KP+ +T ++V +AC+ G GL + + C +V LV
Sbjct: 182 DME-KVGVKPDDITFIAVFTACS-----YSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLV 343
Query: 281 DMYSKCGCWGSAFDVF-----SRSEKRNLITWNSMIAGMMMNSESERAVELFERMV 331
D+ S+ G + A + S + + W + ++ + E++ A E+++
Sbjct: 344 DLLSRAGLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVL 511
>BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis thaliana
DNA chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (14%)
Length = 444
Score = 120 bits (302), Expect = 1e-27
Identities = 65/148 (43%), Positives = 92/148 (61%), Gaps = 1/148 (0%)
Frame = +1
Query: 357 EAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALV 416
EA F MQ G+ P S+++ D V R AK IHG A+R +D + F+ATALV
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 417 DTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDE-MVQP 475
D Y KCG + AR +FD + WNAMI GYGT+G ++A ++F +M +E ++P
Sbjct: 187 DMYAKCGAIETARELFDMMQER--HVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKP 360
Query: 476 NSATFVSVLSACSHSGQIERGLRFFRMI 503
N TF+SV+SACSHSG +E GL +F+++
Sbjct: 361 NDITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 95.1 bits (235), Expect = 7e-20
Identities = 52/132 (39%), Positives = 80/132 (60%), Gaps = 1/132 (0%)
Frame = +1
Query: 235 KPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFD 294
KP+ T VSV++A A LS R K +HGL+++ +V V T+LVDMY+KCG +A +
Sbjct: 49 KPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALVDMYAKCGAIETARE 228
Query: 295 VFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGIL-PDSATWNSLISGFAQKG 353
+F ++R++ITWN+MI G + + A++LF+ M +E L P+ T+ S+IS + G
Sbjct: 229 LFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPNDITFLSVISACSHSG 408
Query: 354 VCVEAFKYFSKM 365
E YF M
Sbjct: 409 FVEEGLYYFKIM 444
Score = 88.2 bits (217), Expect = 9e-18
Identities = 43/131 (32%), Positives = 82/131 (61%), Gaps = 1/131 (0%)
Frame = +1
Query: 120 QAVWLFRQIGFWNIRPNSVTIVSLLSA-RDVKNQSHVQQVHCLACKLGVEYDVYVSTSLV 178
+A+ LF + I+P+S T VS+++A D+ + +H LA + ++ +V+V+T+LV
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 179 TAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNK 238
Y+KCG + ++ ++F+ ++ ++V+T+NA + G +G + D+F DM KPN
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 239 VTLVSVVSACA 249
+T +SV+SAC+
Sbjct: 367 ITFLSVISACS 399
Score = 65.1 bits (157), Expect = 8e-11
Identities = 40/130 (30%), Positives = 71/130 (53%), Gaps = 1/130 (0%)
Frame = +1
Query: 18 EALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALI 77
EALNL+ + S P++FTF ++ A ++LS Q + +H +T ++ +TAL+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 78 ASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQI-GFWNIRPN 136
Y A + A ELFD M + + +NA++ G +G A+ LF + +++PN
Sbjct: 187 DMY-AKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPN 363
Query: 137 SVTIVSLLSA 146
+T +S++SA
Sbjct: 364 DITFLSVISA 393
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 115 bits (289), Expect = 4e-26
Identities = 59/178 (33%), Positives = 108/178 (60%), Gaps = 3/178 (1%)
Frame = +1
Query: 445 WNAMIGGYGTNGDYESAFEVFYEM-LDEMVQPNSATFVSVLSACSHSGQIERGLRFFR-M 502
WN++I G NG AFE F ++ ++++P+S +F+ VL+AC H G I + +F M
Sbjct: 31 WNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKARDYFELM 210
Query: 503 IRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNL 561
+ KY ++P +H+ C+VD+LG+AG L EA +L++ + +P A ++ SLL +CR + + +
Sbjct: 211 MNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCRKHRNVQI 390
Query: 562 GEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
A ++ ++ P + + V++SN++AA ++ E R L+ + +K G S IE+
Sbjct: 391 ARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKEPGCSSIEL 564
Score = 45.1 bits (105), Expect = 8e-05
Identities = 35/145 (24%), Positives = 65/145 (44%), Gaps = 2/145 (1%)
Frame = +1
Query: 205 YNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLS 264
+N+ + GL NG R F+ F + + KP+ V+ + V++AC L I + L
Sbjct: 31 WNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKARDYFELM 210
Query: 265 M-KLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSE-KRNLITWNSMIAGMMMNSESER 322
M K E + T +VD+ + G A ++ K + I W S+++ + +
Sbjct: 211 MNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCRKHRNVQI 390
Query: 323 AVELFERMVDEGILPDSATWNSLIS 347
A +R+ + + P A+ L+S
Sbjct: 391 ARRAAQRVYE--LNPSDASGYVLMS 459
Score = 33.5 bits (75), Expect = 0.25
Identities = 26/110 (23%), Positives = 49/110 (43%), Gaps = 2/110 (1%)
Frame = +1
Query: 342 WNSLISGFAQKGVCVEAFKYFSKMQCAG-VAPCLKILTSLLSVCGDSCVLRSAKAIHGYA 400
WNS+I G A G EAF++FSK++ + + P +L+ C + A+
Sbjct: 31 WNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKARDYFELM 210
Query: 401 L-RICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMI 449
+ + ++ T +VD + G + A + +KP D W +++
Sbjct: 211 MNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKP-DAIIWGSLL 357
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 112 bits (280), Expect = 4e-25
Identities = 57/139 (41%), Positives = 87/139 (62%), Gaps = 2/139 (1%)
Frame = -2
Query: 480 FVSVLSACSHSGQIERGLRFF-RMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL 538
F +VL+AC+H+G I++G +F +MI YGL P + + C++DL R G L +ARDL++E+
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 539 A-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVE 597
+P ++ S L AC+ Y D LG E A++LI +EP N AP + L++IY G W+E
Sbjct: 603 PYDPNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEAS 424
Query: 598 RIRGLITDKGLDKNSGISM 616
+R L+ + K G SM
Sbjct: 423 EVRSLMQQRVKRKPPGWSM 367
Score = 42.0 bits (97), Expect = 7e-04
Identities = 31/133 (23%), Positives = 61/133 (45%), Gaps = 5/133 (3%)
Frame = -2
Query: 175 TSLVTAYSKCGVLVSSNKVFENLRVK-----NVVTYNAFMSGLLQNGFHRVVFDVFKDMT 229
T+++TA + G + + F + ++ Y + +NG R D+ ++M
Sbjct: 780 TAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEMP 601
Query: 230 MNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCW 289
+ PN + S +SAC ++ LG++ +K+E C+ +T L +Y+ G W
Sbjct: 600 YD----PNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLT-LAHIYTTKGLW 436
Query: 290 GSAFDVFSRSEKR 302
A +V S ++R
Sbjct: 435 NEASEVRSLMQQR 397
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 102 bits (254), Expect = 4e-22
Identities = 67/231 (29%), Positives = 121/231 (52%), Gaps = 2/231 (0%)
Frame = +2
Query: 346 ISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICV 405
++ F Q+G EA + K A A C +IL L CG S + AK +H Y L+
Sbjct: 209 LTRFCQEGKVKEALELMEKGIKAD-ANCFEILFDL---CGKSKSVEDAKKVHDYFLQSTF 376
Query: 406 DKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVF 465
D + +++ Y C ++ AR VFD + D W+ MI GY + + ++F
Sbjct: 377 RSDFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDS--WHMMIRGYANSTMGDEGLQLF 550
Query: 466 YEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIR-KYGLDPKPEHFGCVVDLLGR 524
+M + ++ S T ++VLSAC + +E + ++ KYG++P EH+ ++D+LG+
Sbjct: 551 EQMNELGLEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQ 730
Query: 525 AGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEP 574
+G L EA + +++L EP +VF++L R + D +L + + ++ ++P
Sbjct: 731 SGYLKEAEEFIEQLPFEPTVTVFETLKNYARIHGDVDLEDHVEELIVSLDP 883
Score = 58.2 bits (139), Expect = 9e-09
Identities = 36/148 (24%), Positives = 67/148 (44%), Gaps = 1/148 (0%)
Frame = +2
Query: 248 CATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITW 307
C ++ K+VH ++ + +++MY C A VF RN+ +W
Sbjct: 311 CGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSW 490
Query: 308 NSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQC 367
+ MI G ++ + ++LFE+M + G+ S T +++S +A+ Y M+
Sbjct: 491 HMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACGSAEAVEDAYIYLESMKS 670
Query: 368 A-GVAPCLKILTSLLSVCGDSCVLRSAK 394
G+ P ++ LL V G S L+ A+
Sbjct: 671 KYGIEPGVEHYMGLLDVLGQSGYLKEAE 754
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 98.6 bits (244), Expect = 6e-21
Identities = 64/206 (31%), Positives = 113/206 (54%), Gaps = 1/206 (0%)
Frame = +3
Query: 105 FNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA-RDVKNQSHVQQVHCLAC 163
+ A+++G ++NG +AV FR + + N T ++L+A V + +QVH
Sbjct: 93 WTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHGFIV 272
Query: 164 KLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFD 223
K G +VYV ++LV Y+KCG L ++ + E + +VV++N+ M G +++G
Sbjct: 273 KSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALR 452
Query: 224 VFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMY 283
+FK+M K + T SV++ C + +I K VHGL +K ++ +V +LVDMY
Sbjct: 453 LFKNM-HGRNMKIDDYTFPSVLN-CCVVGSIN-PKSVHGLIIKTGFENYKLVSNALVDMY 623
Query: 284 SKCGCWGSAFDVFSRSEKRNLITWNS 309
+K G A+ VF + ++++I+W S
Sbjct: 624 AKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 89.7 bits (221), Expect = 3e-18
Identities = 68/262 (25%), Positives = 121/262 (45%), Gaps = 4/262 (1%)
Frame = +3
Query: 177 LVTAYSKCGVLVSSNKVFENLRV--KNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEE 234
LV Y+KC + + +F+ L KN V + A ++G QNG + F+ M E
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 235 KPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFD 294
N+ T ++++AC+++ G+QVHG +K +V V ++LVDMY+KCG +A +
Sbjct: 183 C-NQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKN 359
Query: 295 VFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGV 354
+ E ++++WNS++ G + + E A+ LF+ M + D T+ S++
Sbjct: 360 MLETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVL-------- 515
Query: 355 CVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRS--AKAIHGYALRICVDKDDFLA 412
+ CV+ S K++HG ++ + ++
Sbjct: 516 -------------------------------NCCVVGSINPKSVHGLIIKTGFENYKLVS 602
Query: 413 TALVDTYMKCGCVSFARFVFDQ 434
ALVD Y K G + A VF++
Sbjct: 603 NALVDMYAKTGDMDCAYTVFEK 668
Score = 85.9 bits (211), Expect = 4e-17
Identities = 55/201 (27%), Positives = 104/201 (51%)
Frame = +3
Query: 7 VTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGF 66
VT NG +A+ + ++H+ N +TFP +L ACS++ + + +H + K+GF
Sbjct: 105 VTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHGFIVKSGF 284
Query: 67 HSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFR 126
S+ + +AL+ Y A A + + M + ++N+++ G R+G +A+ LF+
Sbjct: 285 GSNVYVQSALVDMY-AKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALRLFK 461
Query: 127 QIGFWNIRPNSVTIVSLLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGV 186
+ N++ + T S+L+ V + + VH L K G E VS +LV Y+K G
Sbjct: 462 NMHGRNMKIDDYTFPSVLNC-CVVGSINPKSVHGLIIKTGFENYKLVSNALVDMYAKTGD 638
Query: 187 LVSSNKVFENLRVKNVVTYNA 207
+ + VFE + K+V+++ +
Sbjct: 639 MDCAYTVFEKMLEKDVISWTS 701
Score = 76.6 bits (187), Expect = 3e-14
Identities = 60/262 (22%), Positives = 112/262 (41%)
Frame = +3
Query: 279 LVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPD 338
LVDMY+KC C A +F E F+R +
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLE--------------------------FDRK-------N 83
Query: 339 SATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHG 398
W ++++G+AQ G +A ++F M GV ++L+ C + +HG
Sbjct: 84 HVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHG 263
Query: 399 YALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDY 458
+ ++ + ++ +ALVD Y KCG + A+ + + ++ DD WN+++ G+ +G
Sbjct: 264 FIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLE--TMEDDDVVSWNSLMVGFVRHGLE 437
Query: 459 ESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCV 518
E A +F M ++ + TF SVL+ C + + +I K G + +
Sbjct: 438 EEALRLFKNMHGRNMKIDDYTFPSVLNCCVVGSINPKSVH--GLIIKTGFENYKLVSNAL 611
Query: 519 VDLLGRAGQLGEARDLVQELAE 540
VD+ + G + A + +++ E
Sbjct: 612 VDMYAKTGDMDCAYTVFEKMLE 677
Score = 71.2 bits (173), Expect = 1e-12
Identities = 43/136 (31%), Positives = 66/136 (47%)
Frame = +3
Query: 415 LVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQ 474
LVD Y KC CVS A F+F + + W AM+ GY NGD A E F M + V+
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 475 PNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDL 534
N TF ++L+ACS G + I K G +VD+ + G L A+++
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 535 VQELAEPPASVFDSLL 550
++ + + ++SL+
Sbjct: 363 LETMEDDDVVSWNSLM 410
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 98.6 bits (244), Expect = 6e-21
Identities = 60/229 (26%), Positives = 102/229 (44%)
Frame = +2
Query: 235 KPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFD 294
KPN VT+VSV+ ACA +SN+ G ++H L++ V T+L+DMY KC A D
Sbjct: 38 KPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCFSPEKAVD 217
Query: 295 VFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGV 354
+F+R K+++I W + +G N ++ +F M+ G PD+
Sbjct: 218 IFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIA------------- 358
Query: 355 CVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATA 414
L +L+ + +L+ A H + ++ + + F+ +
Sbjct: 359 ----------------------LVKILTTVSELGILQQAVCFHAFVIKNGFENNQFIGAS 472
Query: 415 LVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFE 463
L++ Y KC + A VF K D W+++I YG +G E A +
Sbjct: 473 LIEVYAKCSSIEDANKVFKGMTYK--DVVTWSSIIAAYGFHGQGEEALK 613
Score = 92.8 bits (229), Expect = 3e-19
Identities = 58/202 (28%), Positives = 104/202 (50%), Gaps = 3/202 (1%)
Frame = +2
Query: 20 LNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIAS 79
L+L S + PN T +L+AC+ +S+ + +H GF STAL+
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 80 YAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVT 139
Y S A+++F+ MP+ + A+ + SG + NG +++W+FR + RP+++
Sbjct: 182 YM-KCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIA 358
Query: 140 IVSLLSARDVKNQSHVQQV---HCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFEN 196
+V +L+ V +QQ H K G E + ++ SL+ Y+KC + +NKVF+
Sbjct: 359 LVKILTT--VSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKG 532
Query: 197 LRVKNVVTYNAFMSGLLQNGFH 218
+ K+VVT+++ ++ GFH
Sbjct: 533 MTYKDVVTWSSIIAAY---GFH 589
Score = 87.0 bits (214), Expect = 2e-17
Identities = 53/194 (27%), Positives = 99/194 (50%), Gaps = 1/194 (0%)
Frame = +2
Query: 133 IRPNSVTIVSLLSA-RDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSN 191
I+PN VT+VS+L A + N ++H LA G E + VST+L+ Y KC +
Sbjct: 35 IKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCFSPEKAV 214
Query: 192 KVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATL 251
+F + K+V+ + SG NG VF++M ++ +P+ + LV +++ + L
Sbjct: 215 DIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNM-LSSGTRPDAIALVKILTTVSEL 391
Query: 252 SNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMI 311
++ H +K ++ + SL+++Y+KC A VF ++++TW+S+I
Sbjct: 392 GILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSII 571
Query: 312 AGMMMNSESERAVE 325
A + + E A++
Sbjct: 572 AAYGFHGQGEEALK 613
Score = 63.9 bits (154), Expect = 2e-10
Identities = 42/190 (22%), Positives = 81/190 (42%)
Frame = +2
Query: 363 SKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKC 422
S+M + P + S+L C L IH A+ + + ++TAL+D YMKC
Sbjct: 14 SEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKC 193
Query: 423 GCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVS 482
A +F++ K D W + GY NG + VF ML +P++ V
Sbjct: 194 FSPEKAVDIFNRMPKK--DVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVK 367
Query: 483 VLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAEPP 542
+L+ S G +++ + F + K G + ++++ + + +A + + +
Sbjct: 368 ILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKD 547
Query: 543 ASVFDSLLGA 552
+ S++ A
Sbjct: 548 VVTWSSIIAA 577
Score = 44.7 bits (104), Expect = 1e-04
Identities = 36/159 (22%), Positives = 65/159 (40%), Gaps = 6/159 (3%)
Frame = +2
Query: 467 EMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAG 526
EMLD+ ++PN T VSVL AC+ +E G++ + YG + + ++D+ +
Sbjct: 17 EMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCF 196
Query: 527 QLGEARDLVQELAEPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAP----LVV 582
+A D+ + + + L Y D+ + E ++ P LV
Sbjct: 197 SPEKAVDIFNRMPKKDVIAWAVLFSG---YADNGMVHESMWVFRNMLSSGTRPDAIALVK 367
Query: 583 LSNIYAALGRWSEVERIRGLITDKGLDKNS--GISMIEV 619
+ + LG + + G + N G S+IEV
Sbjct: 368 ILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEV 484
>BQ138324 weakly similar to GP|12322902|gb| hypothetical protein; 50785-52656
{Arabidopsis thaliana}, partial (7%)
Length = 619
Score = 96.3 bits (238), Expect = 3e-20
Identities = 63/168 (37%), Positives = 94/168 (55%), Gaps = 2/168 (1%)
Frame = +1
Query: 16 YKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTA 75
+ EAL +Y H+ SS PNTFTFP+LLK+C+ LS P LH+H+ KTG P+T ++
Sbjct: 115 FMEALTVYRHMLRSSFFPNTFTFPVLLKSCALLSLPFTGSQLHSHILKTGSQPDPYTHSS 294
Query: 76 LIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWN-IR 134
LI Y + T A ++FDE P ++NA++SG + N +A+ LFR++ N
Sbjct: 295 LINMY-SKTSLPCLARKVFDESPVNLTISYNAMISGYTNNMMIVEAIKLFRRMLCENRFF 471
Query: 135 PNSVTIVSLLSARDVKNQSHVQ-QVHCLACKLGVEYDVYVSTSLVTAY 181
NSVT++ L+S V + + +H K G E D+ V +T Y
Sbjct: 472 VNSVTMLGLVSGILVPEKLRLGFCLHGCCFKFGFENDLXVGNXFLTMY 615
Score = 68.6 bits (166), Expect = 7e-12
Identities = 52/189 (27%), Positives = 84/189 (43%), Gaps = 1/189 (0%)
Frame = +1
Query: 96 EMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARDVKNQSHV 155
E Q TA+N L LS+ +A+ ++R + + PN+ T LL + + +
Sbjct: 49 ESKQNPTTAWNCYLRELSKQRKFMEALTVYRHMLRSSFFPNTFTFPVLLKSCALLSLPFT 228
Query: 156 -QQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQ 214
Q+H K G + D Y +SL+ YSK + + KVF+ V ++YNA +SG
Sbjct: 229 GSQLHSHILKTGSQPDPYTHSSLINMYSKTSLPCLARKVFDESPVNLTISYNAMISGYTN 408
Query: 215 NGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVM 274
N +F+ M N VT++ +VS +RLG +HG K + +
Sbjct: 409 NMMIVEAIKLFRRMLCENRFFVNSVTMLGLVSGILVPEKLRLGFCLHGCCFKFGFENDLX 588
Query: 275 VVTSLVDMY 283
V + MY
Sbjct: 589 VGNXFLTMY 615
Score = 54.3 bits (129), Expect = 1e-07
Identities = 38/114 (33%), Positives = 56/114 (48%), Gaps = 1/114 (0%)
Frame = +1
Query: 236 PNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDV 295
PN T ++ +CA LS G Q+H +K + +SL++MYSK A V
Sbjct: 166 PNTFTFPVLLKSCALLSLPFTGSQLHSHILKTGSQPDPYTHSSLINMYSKTSLPCLARKV 345
Query: 296 FSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEG-ILPDSATWNSLISG 348
F S I++N+MI+G N A++LF RM+ E +S T L+SG
Sbjct: 346 FDESPVNLTISYNAMISGYTNNMMIVEAIKLFRRMLCENRFFVNSVTMLGLVSG 507
Score = 46.6 bits (109), Expect = 3e-05
Identities = 39/172 (22%), Positives = 69/172 (39%), Gaps = 1/172 (0%)
Frame = +1
Query: 339 SATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHG 398
+ WN + +++ +EA + M + P LL C + + +H
Sbjct: 67 TTAWNCYLRELSKQRKFMEALTVYRHMLRSSFFPNTFTFPVLLKSCALLSLPFTGSQLHS 246
Query: 399 YALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDY 458
+ L+ D + ++L++ Y K AR VFD+ V +NAMI GY N
Sbjct: 247 HILKTGSQPDPYTHSSLINMYSKTSLPCLARKVFDESPVNLT--ISYNAMISGYTNNMMI 420
Query: 459 ESAFEVFYEML-DEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLD 509
A ++F ML + NS T + ++S ++ G K+G +
Sbjct: 421 VEAIKLFRRMLCENRFFVNSVTMLGLVSGILVPEKLRLGFCLHGCCFKFGFE 576
Score = 31.2 bits (69), Expect = 1.2
Identities = 22/100 (22%), Positives = 37/100 (37%)
Frame = +1
Query: 438 KPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGL 497
K + WN + + A V+ ML PN+ TF +L +C+ G
Sbjct: 55 KQNPTTAWNCYLRELSKQRKFMEALTVYRHMLRSSFFPNTFTFPVLLKSCALLSLPFTGS 234
Query: 498 RFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQE 537
+ I K G P P ++++ + AR + E
Sbjct: 235 QLHSHILKTGSQPDPYTHSSLINMYSKTSLPCLARKVFDE 354
>BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T6L1.11
[imported] - Arabidopsis thaliana, partial (20%)
Length = 461
Score = 96.3 bits (238), Expect = 3e-20
Identities = 53/149 (35%), Positives = 87/149 (57%), Gaps = 1/149 (0%)
Frame = +3
Query: 203 VTYNAFMSGLLQNGFHRVVFDVFKDMTM-NLEEKPNKVTLVSVVSACATLSNIRLGKQVH 261
+++ + ++G QNG R D+F++M + NL+ ++ T SV++AC + ++ GKQVH
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQM--DQYTFGSVLTACGGVMALQEGKQVH 179
Query: 262 GLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESE 321
++ + D++ V ++LVDMY KC SA VF + +N+++W +M+ G N SE
Sbjct: 180 AYIIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSE 359
Query: 322 RAVELFERMVDEGILPDSATWNSLISGFA 350
AV+ F M GI PD T S+IS A
Sbjct: 360 EAVKTFSDMQKYGIEPDDFTLGSVISSCA 446
Score = 93.2 bits (230), Expect = 3e-19
Identities = 51/151 (33%), Positives = 80/151 (52%)
Frame = +3
Query: 339 SATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHG 398
S +W S+I+GF Q G+ +A F +M+ + S+L+ CG L+ K +H
Sbjct: 3 SISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHA 182
Query: 399 YALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDY 458
Y +R + F+A+ALVD Y KC + A VF + K + W AM+ GYG NG
Sbjct: 183 YIIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCK--NVVSWTAMLVGYGQNGYS 356
Query: 459 ESAFEVFYEMLDEMVQPNSATFVSVLSACSH 489
E A + F +M ++P+ T SV+S+C++
Sbjct: 357 EEAVKTFSDMQKYGIEPDDFTLGSVISSCAN 449
Score = 78.2 bits (191), Expect = 9e-15
Identities = 45/152 (29%), Positives = 86/152 (55%), Gaps = 1/152 (0%)
Frame = +3
Query: 104 AFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA-RDVKNQSHVQQVHCLA 162
++ ++++G ++NG A+ +FR++ N++ + T S+L+A V +QVH
Sbjct: 9 SWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAYI 188
Query: 163 CKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVF 222
+ + +++V+++LV Y KC + S+ VF+ + KNVV++ A + G QNG+
Sbjct: 189 IRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAV 368
Query: 223 DVFKDMTMNLEEKPNKVTLVSVVSACATLSNI 254
F DM +P+ TL SV+S+CA L+++
Sbjct: 369 KTFSDM-QKYGIEPDDFTLGSVISSCANLASL 461
Score = 59.7 bits (143), Expect = 3e-09
Identities = 32/140 (22%), Positives = 74/140 (52%)
Frame = +3
Query: 7 VTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGF 66
+T NGL ++A++++ + + + +TF +L AC + + + + +HA++ +T +
Sbjct: 24 ITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAYIIRTDY 203
Query: 67 HSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFR 126
+ ++AL+ Y ++ A +F +M + ++ A+L G +NG +AV F
Sbjct: 204 KDNIFVASALVDMY-CKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAVKTFS 380
Query: 127 QIGFWNIRPNSVTIVSLLSA 146
+ + I P+ T+ S++S+
Sbjct: 381 DMQKYGIEPDDFTLGSVISS 440
Score = 38.9 bits (89), Expect = 0.006
Identities = 20/52 (38%), Positives = 29/52 (55%)
Frame = +3
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERG 496
W +MI G+ NG A ++F EM E +Q + TF SVL+AC ++ G
Sbjct: 12 WTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEG 167
>TC83657 weakly similar to GP|11994382|dbj|BAB02341.
gb|AAB82628.1~gene_id:MIL23.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1299
Score = 95.5 bits (236), Expect = 5e-20
Identities = 54/140 (38%), Positives = 84/140 (59%), Gaps = 3/140 (2%)
Frame = +3
Query: 481 VSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL-A 539
+ VLSAC+H G + L + +YG++ H+GC+VDLLGRAG+L EA +L++ +
Sbjct: 78 LXVLSACAHGGLMSEALEVISKMEEYGIEMGIRHYGCMVDLLGRAGKLKEAYELIKRMPM 257
Query: 540 EPPASVFDSLLGACRCYLDSNLGEEMAMKLI--DIEPKNPAPLVVLSNIYAALGRWSEVE 597
+P +V +++GAC + D + E++ MK+I D + V+LSNIYAA +W + E
Sbjct: 258 KPNETVLGAMIGACWIHSDMKMAEQV-MKMIGADSAACVNSHNVLLSNIYAASEKWEKAE 434
Query: 598 RIRGLITDKGLDKNSGISMI 617
IR + D G +K G S I
Sbjct: 435 MIRSSMVDGGSEKIPGYSSI 494
Score = 32.3 bits (72), Expect = 0.55
Identities = 32/138 (23%), Positives = 60/138 (43%), Gaps = 11/138 (7%)
Frame = +3
Query: 166 GVEYDVYVSTSLVTAYSKC---GVLVSSNKVFENLRVKNVVT----YNAFMSGLLQNGFH 218
GV+Y + L+ S C G++ + +V + + Y + L + G
Sbjct: 42 GVKYKA*CXSRLLXVLSACAHGGLMSEALEVISKMEEYGIEMGIRHYGCMVDLLGRAGKL 221
Query: 219 RVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTS 278
+ +++ K M M KPN+ L +++ AC S++++ +QV MK+ D V S
Sbjct: 222 KEAYELIKRMPM----KPNETVLGAMIGACWIHSDMKMAEQV----MKMIGADSAACVNS 377
Query: 279 ----LVDMYSKCGCWGSA 292
L ++Y+ W A
Sbjct: 378 HNVLLSNIYAASEKWEKA 431
>TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein At2g34400
[imported] - Arabidopsis thaliana, partial (5%)
Length = 647
Score = 93.2 bits (230), Expect = 3e-19
Identities = 52/203 (25%), Positives = 105/203 (51%), Gaps = 1/203 (0%)
Frame = +1
Query: 67 HSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFR 126
HSH S ++ + N FHY+L +F+ P++ +N ++ + + LF
Sbjct: 4 HSHKLFSVSI---HLNNNNYFHYSLSIFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFN 174
Query: 127 QIGFWNIRPNSVTIVSLLSA-RDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCG 185
Q+ + P++ T +L A + + ++H K G++ D YVS S + Y++ G
Sbjct: 175 QLRLNGLYPDNYTYPFVLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELG 354
Query: 186 VLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVV 245
+ K+F+ + ++ V++N +SG ++ +VF+ M ++ EK ++ T+VS +
Sbjct: 355 RIDFVRKLFDEISERDSVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSL 534
Query: 246 SACATLSNIRLGKQVHGLSMKLE 268
+ACA N+ +GK++HG ++ E
Sbjct: 535 TACAASRNVEVGKEIHGFIIEKE 603
Score = 68.6 bits (166), Expect = 7e-12
Identities = 47/174 (27%), Positives = 77/174 (44%), Gaps = 1/174 (0%)
Frame = +1
Query: 337 PDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAI 396
P +N LI F ++ F++++ G+ P +L R I
Sbjct: 94 PSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIADFRQGTKI 273
Query: 397 HGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNG 456
H + + +D D +++ + +D Y + G + F R +FD+ + D WN MI G
Sbjct: 274 HAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISER--DSVSWNVMISGCVKCR 447
Query: 457 DYESAFEVFYEM-LDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLD 509
+E A EVF M +D + + AT VS L+AC+ S +E G I + LD
Sbjct: 448 RFEEAVEVFQRMRVDSNEKISEATVVSSLTACAASRNVEVGKEIHGFIIEKELD 609
Score = 59.3 bits (142), Expect = 4e-09
Identities = 35/141 (24%), Positives = 65/141 (45%)
Frame = +1
Query: 190 SNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACA 249
S +F + ++ YN + + + + +F + +N P+ T V+ A A
Sbjct: 64 SLSIFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLN-GLYPDNYTYPFVLKAVA 240
Query: 250 TLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNS 309
+++ R G ++H K V S +DMY++ G +F +R+ ++WN
Sbjct: 241 FIADFRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISERDSVSWNV 420
Query: 310 MIAGMMMNSESERAVELFERM 330
MI+G + E AVE+F+RM
Sbjct: 421 MISGCVKCRRFEEAVEVFQRM 483
Score = 56.6 bits (135), Expect = 3e-08
Identities = 36/132 (27%), Positives = 73/132 (55%), Gaps = 1/132 (0%)
Frame = +1
Query: 16 YKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTA 75
++ ++L++ L + P+ +T+P +LKA + ++ Q +HA +FKTG S + S +
Sbjct: 148 FQTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNS 327
Query: 76 LIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGF-WNIR 134
+ YA R + +LFDE+ + ++N ++SG + +AV +F+++ N +
Sbjct: 328 FMDMYAELGR-IDFVRKLFDEISERDSVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEK 504
Query: 135 PNSVTIVSLLSA 146
+ T+VS L+A
Sbjct: 505 ISEATVVSSLTA 540
>BG646586 weakly similar to GP|10177683|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (9%)
Length = 687
Score = 89.4 bits (220), Expect = 4e-18
Identities = 47/112 (41%), Positives = 71/112 (62%), Gaps = 1/112 (0%)
Frame = -3
Query: 508 LDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLGEEMA 566
++P PEH+ +VDLLGRAGQL +A L++E + A+ + SLL ACR Y + LGE A
Sbjct: 664 IEPLPEHYTGMVDLLGRAGQLEKAYSLIKENSTSADATTWGSLLAACRVYGNVELGEIAA 485
Query: 567 MKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIE 618
L +I+P + V+L+N YA+ +W E ++ L++ KG+ K SG S I+
Sbjct: 484 RHLFEIDPTDSGNYVLLANTYASNDKWERAEEVKKLMSKKGMKKPSGYSWIQ 329
>AW687696 weakly similar to PIR|T49965|T499 hypothetical protein F8M21.190 -
Arabidopsis thaliana, partial (16%)
Length = 289
Score = 80.9 bits (198), Expect = 1e-15
Identities = 41/96 (42%), Positives = 63/96 (64%), Gaps = 2/96 (2%)
Frame = +2
Query: 470 DEMVQPNSATFVSVLSACSHSGQIERGLRFFRMI-RKYGLDPKPEHFGCVVDLLGRAGQL 528
+ + +P+ TF+ VL ACSH G ++ G R+F ++ R Y + P +H+GC+VDLLGRAG
Sbjct: 2 ENVERPDEITFLCVLCACSHGGLVDEGRRYFEIMNRDYNIKPTIKHYGCMVDLLGRAGLF 181
Query: 529 GEARDLVQEL-AEPPASVFDSLLGACRCYLDSNLGE 563
EA +L++ + E A ++ +LL ACR Y + LGE
Sbjct: 182 VEAYELIKSMPVECNAIIWRTLLAACRNYGNVELGE 289
>BE320932 weakly similar to PIR|T04548|T045 hypothetical protein F28J12.180 -
Arabidopsis thaliana, partial (14%)
Length = 354
Score = 79.7 bits (195), Expect = 3e-15
Identities = 40/105 (38%), Positives = 64/105 (60%)
Frame = +3
Query: 157 QVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNG 216
Q+H K + DV++ TSL+ Y+KCG +VSS KVF+ ++V+N T+ + +SG +NG
Sbjct: 39 QLHGAIVKKICKSDVFIGTSLIDMYAKCGEIVSSKKVFDRMKVRNTATWTSIISGYARNG 218
Query: 217 FHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVH 261
F + F+ M + NK TLV V++AC T+ +G++VH
Sbjct: 219 FGEEALNFFRLMKRK-KVYVNKSTLVCVMTACGTIKASLIGREVH 350
Score = 63.2 bits (152), Expect = 3e-10
Identities = 33/92 (35%), Positives = 51/92 (54%)
Frame = +3
Query: 396 IHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTN 455
+HG ++ D F+ T+L+D Y KCG + ++ VFD+ V+ + A W ++I GY N
Sbjct: 42 LHGAIVKKICKSDVFIGTSLIDMYAKCGEIVSSKKVFDRMKVR--NTATWTSIISGYARN 215
Query: 456 GDYESAFEVFYEMLDEMVQPNSATFVSVLSAC 487
G E A F M + V N +T V V++AC
Sbjct: 216 GFGEEALNFFRLMKRKKVYVNKSTLVCVMTAC 311
Score = 60.5 bits (145), Expect = 2e-09
Identities = 32/87 (36%), Positives = 47/87 (53%)
Frame = +3
Query: 255 RLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGM 314
+ G Q+HG +K V + TSL+DMY+KCG S+ VF R + RN TW S+I+G
Sbjct: 27 KCGTQLHGAIVKKICKSDVFIGTSLIDMYAKCGEIVSSKKVFDRMKVRNTATWTSIISGY 206
Query: 315 MMNSESERAVELFERMVDEGILPDSAT 341
N E A+ F M + + + +T
Sbjct: 207 ARNGFGEEALNFFRLMKRKKVYVNKST 287
Score = 32.7 bits (73), Expect = 0.42
Identities = 24/90 (26%), Positives = 42/90 (46%)
Frame = +3
Query: 57 LHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNG 116
LH + K S T+LI YA + ++FD M + +++SG +RNG
Sbjct: 42 LHGAIVKKICKSDVFIGTSLIDMYA-KCGEIVSSKKVFDRMKVRNTATWTSIISGYARNG 218
Query: 117 PRGQAVWLFRQIGFWNIRPNSVTIVSLLSA 146
+A+ FR + + N T+V +++A
Sbjct: 219 FGEEALNFFRLMKRKKVYVNKSTLVCVMTA 308
>BF635667 weakly similar to PIR|C85359|C853 hypothetical protein AT4g30700
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 79.0 bits (193), Expect = 5e-15
Identities = 48/140 (34%), Positives = 73/140 (51%), Gaps = 3/140 (2%)
Frame = +3
Query: 483 VLSACSHSGQIERGLRFF-RMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL--A 539
V++ C G +++G + F MI YGL+P EH+ C+VDL R G+L EA L++ + +
Sbjct: 15 VVATC---GLLDQGKKIFGSMISDYGLEPTAEHYACMVDLFSRCGRLEEALQLLERMKSS 185
Query: 540 EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERI 599
S++ +LL C + + +GE A L +EP N + V L IY + G V I
Sbjct: 186 SVTGSMWGALLAGCVMHQNVKIGEVAAHHLFQLEPNNTSNYVALWGIYQSRGMVLGVSTI 365
Query: 600 RGLITDKGLDKNSGISMIEV 619
G + D GL K G I +
Sbjct: 366 XGKMRDLGLVKTPGCXWINI 425
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,837,029
Number of Sequences: 36976
Number of extensions: 302422
Number of successful extensions: 1985
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 1667
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1887
length of query: 620
length of database: 9,014,727
effective HSP length: 102
effective length of query: 518
effective length of database: 5,243,175
effective search space: 2715964650
effective search space used: 2715964650
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136286.11


