

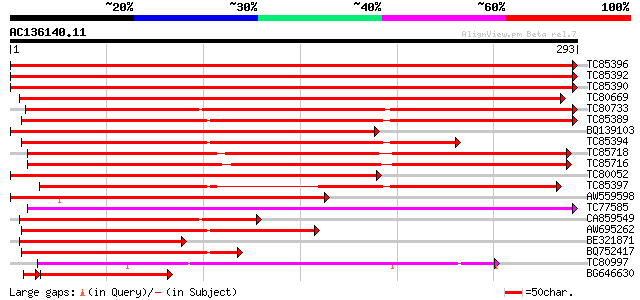
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.11 - phase: 0 /pseudo
(293 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 565 e-161
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 550 e-157
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 549 e-157
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 507 e-144
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 458 e-129
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 377 e-105
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 366 e-102
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 313 7e-86
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 295 2e-80
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 293 7e-80
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 276 9e-75
TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated... 267 3e-72
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 217 4e-57
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 206 1e-53
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 202 1e-52
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 191 4e-49
BE321871 GP|20563125|db heat shock cognate protein {Bombyx mori}... 152 1e-37
BQ752417 similar to GP|5597020|dbj ER chaperone BiP {Aspergillus... 147 5e-36
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 122 2e-28
BG646630 homologue to GP|21479|emb|CAA 70-Kd heat shock protein... 118 9e-28
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 565 bits (1455), Expect = e-161
Identities = 286/293 (97%), Positives = 289/293 (98%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 286
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGE+K FA+
Sbjct: 287 QVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLFAS 466
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEAYLG T+KNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP
Sbjct: 467 EEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 646
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 647 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 826
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMV HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 827 NRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 985
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 550 bits (1417), Expect = e-157
Identities = 275/293 (93%), Positives = 286/293 (96%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 309
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRRISD SVQSDMKLWPFKVI GP +KPMI VNYK E+KQF+A
Sbjct: 310 QVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 489
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+EIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGVI+GLNV+RIINEP
Sbjct: 490 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 669
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 670 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 849
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISGNPRALRR+RTACERAKRTLSSTAQTTIEIDSL
Sbjct: 850 NRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSL 1008
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 549 bits (1414), Expect = e-157
Identities = 275/293 (93%), Positives = 284/293 (96%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 315
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRR D SVQSDMKLWPFKVI GP +KPMI VNYK E+KQF+A
Sbjct: 316 QVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 495
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+EIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGVI+GLNVLRIINEP
Sbjct: 496 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEP 675
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 676 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 855
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 856 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 1014
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 507 bits (1305), Expect = e-144
Identities = 255/282 (90%), Positives = 267/282 (94%)
Frame = +2
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
EG AIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 110 EGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 289
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISS 125
P NTVFDAKRLIGRR SD SVQ+DMKLWPFKV+ GP EKPMI VNYKGE+K+FAAEEISS
Sbjct: 290 PQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAEEISS 469
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
MVLIKMRE+AEA+LG VKNAVVTVPAYFNDSQRQATKDAG I+GLNVLRIINEPTAAAI
Sbjct: 470 MVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 649
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKKA+ GE+NVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN
Sbjct: 650 AYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 829
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTT 287
HFV EF+RKNKKDISGN RALRRLRTACERAKRTLSSTAQTT
Sbjct: 830 HFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 458 bits (1178), Expect = e-129
Identities = 236/285 (82%), Positives = 258/285 (89%)
Frame = +2
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVN 68
AIGIDLGTTYSCVG + +D++EIIANDQGNRTTPSYVAF D+ERLIGDAAKNQVAMNP N
Sbjct: 188 AIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNPHN 367
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
TVFDAKRLIGR+ SD+ VQ+DMK +PFKVI G KP I V +KGE+K F EEIS+MVL
Sbjct: 368 TVFDAKRLIGRKFSDSEVQADMKHFPFKVI-DKGGKPNIEVEFKGENKTFTPEEISAMVL 544
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
+KMRE AEAYLG V NAV+TVPAYFNDSQRQATKDAG+IAGLNVLRIINEPTAAAIAYG
Sbjct: 545 VKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIAYG 724
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
LDKKA GE+NVLIFDLGGGTFDVSLLTIEEGIFEVK+TAGDTHLGGEDFDNR+VNHFV
Sbjct: 725 LDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNHFV 898
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
EFKRK+KKD+S N RALRRLRTACERAKRTLSS+AQT+IEIDSL
Sbjct: 899 NEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSL 1033
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 377 bits (967), Expect = e-105
Identities = 193/288 (67%), Positives = 233/288 (80%), Gaps = 1/288 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP
Sbjct: 155 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNP 334
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ +D VQ DMKL P+K++ G KP I V K GE K F+ EE+S+
Sbjct: 335 ERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKDG-KPYIQVRVKDGETKVFSPEEVSA 511
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEA+LG T+++AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 512 MILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 691
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV +T GDTHLGGEDFD R++
Sbjct: 692 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRIME 862
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ K+K+ KDIS + RAL +LR ERAKR LSS Q +EI+SL
Sbjct: 863 YFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESL 1006
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 366 bits (939), Expect = e-102
Identities = 180/191 (94%), Positives = 188/191 (98%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 235
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR SDASVQSDMKLWPFK+I+GP EKP+IGVNYKGEDK+FAA
Sbjct: 236 QVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEFAA 415
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KMREIAEAYLGS +KNAVVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEP
Sbjct: 416 EEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 595
Query: 181 TAAAIAYGLDK 191
TAAAIAYGLDK
Sbjct: 596 TAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 313 bits (801), Expect = 7e-86
Identities = 161/228 (70%), Positives = 187/228 (81%), Gaps = 1/228 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP
Sbjct: 289 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNP 468
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ D VQ DMKL P+K++ G KP I V K GE K F+ EEIS+
Sbjct: 469 ERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNRDG-KPYIQVRVKDGETKVFSPEEISA 645
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AE +LG T+++AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 646 MILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 825
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTH 233
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTH
Sbjct: 826 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 295 bits (754), Expect = 2e-80
Identities = 156/282 (55%), Positives = 196/282 (69%), Gaps = 1/282 (0%)
Frame = +2
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 442
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T+ AKRLIGRR D Q +MK+ P+K++ P + + + +Q++ +I + VL
Sbjct: 443 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAFVL 610
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG TV AV+TVPAYFND+QRQATKDAG IAGL VLRIINEPTAAA++YG
Sbjct: 611 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 790
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 791 MNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 952
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
EFKR D+S + AL+RLR A E+AK LSST+QT I +
Sbjct: 953 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINL 1078
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 293 bits (749), Expect = 7e-80
Identities = 154/282 (54%), Positives = 195/282 (68%), Gaps = 1/282 (0%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV + + ++I N +G RTTPS VAF E L+G AK Q NP N
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNPTN 426
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T+F KRLIGRR D Q +MK+ P+K++ P + +N +Q++ +I + VL
Sbjct: 427 TLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAFVL 594
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG T+ AVVTVPAYFND+QRQATKDAG IAGL V RIINEPTAAA++YG
Sbjct: 595 TKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALSYG 774
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++ K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 775 MNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 939
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
EFKR + D++ + AL+RLR A E+AK LSST+QT I +
Sbjct: 940 SEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINL 1065
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 276 bits (705), Expect = 9e-75
Identities = 137/192 (71%), Positives = 160/192 (82%)
Frame = +3
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M+G+G+ AIGIDLGTTYSCV V ++ ++II ND G RTTPS+VAF DSER+IGDAA N
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFN 260
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
A NP NT+FDAKRLIGR+ SD VQSD+KLWPFKVI +KPMI VNY E+K FAA
Sbjct: 261 IAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAA 440
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KMREIAE +LGS V++ V+TVPAYFNDSQRQ+T+DAG IAGLNV+RIINEP
Sbjct: 441 EEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEP 620
Query: 181 TAAAIAYGLDKK 192
TAAAIAYG + K
Sbjct: 621 TAAAIAYGFNTK 656
>TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated protein 78
{Lycopersicon esculentum}, partial (57%)
Length = 648
Score = 267 bits (683), Expect = 3e-72
Identities = 150/270 (55%), Positives = 175/270 (64%)
Frame = +1
Query: 16 TTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFDAKR 75
TTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP T+FD KR
Sbjct: 1 TTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNPERTIFDVKR 180
Query: 76 LIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIA 135
LIGR+ D VQ DMKL P+K++ G KP I
Sbjct: 181 LIGRKFEDKEVQRDMKLVPYKIVNRDG-KPYI---------------------------- 273
Query: 136 EAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDKKATS 195
QATKDAGVIAGLNV RIINEPTAAAIAYGLDKK
Sbjct: 274 ------------------------QATKDAGVIAGLNVARIINEPTAAAIAYGLDKKG-- 375
Query: 196 VGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKN 255
GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTHLGGEDFD R+ +F++ K+K+
Sbjct: 376 -GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIREYFIKLIKKKH 552
Query: 256 KKDISGNPRALRRLRTACERAKRTLSSTAQ 285
KDIS + RAL +LR ERAKR LSS Q
Sbjct: 553 SKDISKDNRALGKLRKESERAKRALSSQHQ 642
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 217 bits (553), Expect = 4e-57
Identities = 109/167 (65%), Positives = 133/167 (79%), Gaps = 2/167 (1%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
MA E A+GIDLGTTYSCV VW + +RVEII ND+G++ TPS+VAFTD +RL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 59 KNQVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQF 118
K+Q A+NP NTVFDAKRLIGR+ SD VQ D+ LWPFKV++G +KPMI + +KG++K
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 119 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDA 165
AEEISS+VL MREIAE YL S VKNA +TVPAYFND+QR+AT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 206 bits (523), Expect = 1e-53
Identities = 106/285 (37%), Positives = 160/285 (55%), Gaps = 1/285 (0%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNT 69
+G D G V V + ++++ ND+ R TP+ V F D +R IG A MNP N+
Sbjct: 247 VGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 426
Query: 70 VFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLI 129
+ KRLIG++ +D +Q D+K PF V GP P+I Y GE ++F A ++ M+L
Sbjct: 427 ISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMMLS 606
Query: 130 KMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGL 189
++EIA+ L + V + + +P YF D QR++ DA IAGL+ L +I+E TA A+AYG+
Sbjct: 607 NLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAYGI 786
Query: 190 DKKATSVGE-KNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
K E NV D+G + V + ++G V + + D LGG DFD + +HF
Sbjct: 787 YKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHHFA 966
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+FK + K D+ N RA RLR ACE+ K+ LS+ + + I+ L
Sbjct: 967 AKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECL 1101
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 202 bits (515), Expect = 1e-52
Identities = 103/125 (82%), Positives = 107/125 (85%)
Frame = +3
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
EG A+GIDLGTTYSCVGVWQ+DRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 156 EGKAVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 335
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISS 125
P NTVFDAKRLIGRR +D VQSDMK WPFKVI KP I V YKGE KQF EEISS
Sbjct: 336 PYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVI-DKAAKPYIQVEYKGETKQFTPEEISS 512
Query: 126 MVLIK 130
MVL K
Sbjct: 513 MVLTK 527
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 191 bits (484), Expect = 4e-49
Identities = 99/156 (63%), Positives = 122/156 (77%), Gaps = 2/156 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP
Sbjct: 128 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNP 307
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ +D VQ DMKL P+K++ G KP I V K GE K F+ EE+S+
Sbjct: 308 ERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKDG-KPYIQVRVKDGETKVFSPEEVSA 484
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYF-NDSQRQ 160
M+L KM+E AEA+LG T+++AVVTVPA ND+QRQ
Sbjct: 485 MILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
>BE321871 GP|20563125|db heat shock cognate protein {Bombyx mori}, partial
(13%)
Length = 346
Score = 152 bits (384), Expect = 1e-37
Identities = 72/86 (83%), Positives = 81/86 (93%)
Frame = +1
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
+ PA+GIDLGTTYSCVGV+QH +VEIIANDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 85 KAPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 264
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMK 91
P NT+FDAKRLIGR+ DA+VQ+DMK
Sbjct: 265 PNNTIFDAKRLIGRKFEDATVQADMK 342
>BQ752417 similar to GP|5597020|dbj ER chaperone BiP {Aspergillus oryzae},
partial (18%)
Length = 690
Score = 147 bits (371), Expect = 5e-36
Identities = 71/114 (62%), Positives = 90/114 (78%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV Q +VEI+ NDQGNR TPSYVAFT+ ERL+GDAAKNQ A NP
Sbjct: 322 GTVIGIDLGTTYSCVGVMQGGKVEILVNDQGNRITPSYVAFTEEERLVGDAAKNQAAANP 501
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
NT+FD KR+IG++ +D +VQSD+K +P+KVI G KP++ V+ +G K+F +
Sbjct: 502 YNTIFDIKRMIGQKFADKAVQSDVKHFPYKVIEKDG-KPVVEVDVQGSAKRFTS 660
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 122 bits (305), Expect = 2e-28
Identities = 84/252 (33%), Positives = 131/252 (51%), Gaps = 13/252 (5%)
Frame = +3
Query: 15 GTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNPVNTVFD 72
GT+ V VW VE++ N + + S+V F D G ++ ++ M +T+F+
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 182
Query: 73 AKRLIGRRISDASVQSDMKLWPFKV-IAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKM 131
KRLIGR +D V + L PF V G +P I + EE+ +M L+++
Sbjct: 183 MKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVEL 359
Query: 132 REIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDK 191
R + E +L ++N V+TVP F+ Q + A +AGL+VLR++ EPTA A+ YG +
Sbjct: 360 RLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQ 539
Query: 192 KATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
+ S EK LIF++G G DV++ G+ ++KA AG T +GGED M+
Sbjct: 540 QKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNMMR 716
Query: 246 HFVQE----FKR 253
H + + FKR
Sbjct: 717 HLLPDSENIFKR 752
>BG646630 homologue to GP|21479|emb|CAA 70-Kd heat shock protein {Solanum
tuberosum}, partial (90%)
Length = 526
Score = 118 bits (296), Expect(2) = 9e-28
Identities = 58/68 (85%), Positives = 60/68 (87%)
Frame = +3
Query: 17 TYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFDAKRL 76
TYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVF +
Sbjct: 30 TYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFGEISI 209
Query: 77 IGRRISDA 84
+ RI A
Sbjct: 210LNIRIIKA 233
Score = 22.3 bits (46), Expect(2) = 9e-28
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 8 PAIGIDLGT 16
PAIGIDLGT
Sbjct: 2 PAIGIDLGT 28
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,245,529
Number of Sequences: 36976
Number of extensions: 62956
Number of successful extensions: 324
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 304
length of query: 293
length of database: 9,014,727
effective HSP length: 95
effective length of query: 198
effective length of database: 5,502,007
effective search space: 1089397386
effective search space used: 1089397386
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136140.11


