

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135848.10 - phase: 0
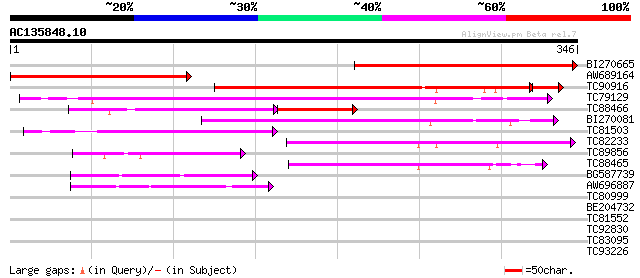
(346 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI270665 similar to GP|22091622|emb cyclin D2 {Daucus carota}, p... 267 5e-72
AW689164 weakly similar to GP|19070617|gb| D-type cyclin {Zea ma... 230 5e-61
TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red... 193 4e-51
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 180 6e-46
TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersic... 92 9e-30
BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}... 123 1e-28
TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhi... 95 5e-20
TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalf... 75 4e-14
TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medica... 66 2e-11
TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersic... 65 4e-11
BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%) 60 9e-10
AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 57 8e-09
TC80999 similar to GP|10176986|dbj|BAB10218. gene_id:MYH19.13~pi... 30 1.8
BE204732 weakly similar to GP|8978275|db receptor protein kinase... 29 3.0
TC81552 similar to GP|16755592|gb|AAL28022.1 small GTPase Rab2 {... 28 4.0
TC92830 weakly similar to PIR|A85043|A85043 probable LRR recepto... 28 5.2
TC83095 similar to GP|9293910|dbj|BAB01813.1 gene_id:MXO21.1~unk... 28 6.8
TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein... 27 8.9
>BI270665 similar to GP|22091622|emb cyclin D2 {Daucus carota}, partial (16%)
Length = 649
Score = 267 bits (682), Expect = 5e-72
Identities = 136/136 (100%), Positives = 136/136 (100%)
Frame = +3
Query: 211 KYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVAISLKELPTQEVDKAITDFFIVDK 270
KYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVAISLKELPTQEVDKAITDFFIVDK
Sbjct: 3 KYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVAISLKELPTQEVDKAITDFFIVDK 182
Query: 271 ERVLKCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSP 330
ERVLKCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSP
Sbjct: 183 ERVLKCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSP 362
Query: 331 NAKRMKFDGPSSGTSQ 346
NAKRMKFDGPSSGTSQ
Sbjct: 363 NAKRMKFDGPSSGTSQ 410
>AW689164 weakly similar to GP|19070617|gb| D-type cyclin {Zea mays}, partial
(27%)
Length = 463
Score = 230 bits (587), Expect = 5e-61
Identities = 110/111 (99%), Positives = 110/111 (99%)
Frame = +1
Query: 1 MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE 60
MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE
Sbjct: 130 MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE 309
Query: 61 IVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPL 111
IVKVMVEKEKDHLPREDYLIRLRGGDLDLSV REALDWIWKAHAYYGFGPL
Sbjct: 310 IVKVMVEKEKDHLPREDYLIRLRGGDLDLSVTREALDWIWKAHAYYGFGPL 462
>TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red goosefoot,
partial (31%)
Length = 886
Score = 193 bits (490), Expect(2) = 4e-51
Identities = 113/204 (55%), Positives = 149/204 (72%), Gaps = 9/204 (4%)
Frame = +3
Query: 126 VFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILS 185
V++FP+G WT+QLLAVAC SLAAK+EE VPQ +DLQ+GE KFVF+AKTIQRMEL++LS
Sbjct: 3 VYEFPKGRAWTMQLLAVACLSLAAKVEETAVPQPLDLQIGESKFVFEAKTIQRMELLVLS 182
Query: 186 SLGWKMRALTPCSFIDYFLAKI-SCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAA 244
+L W+M+A+TP SFI+ FL+KI +K S I+RS QLI + IKG+DFLEF+ SEIAA
Sbjct: 183 TLKWRMQAITPFSFIECFLSKIKDDDKSSLSSSISRSTQLISSTIKGLDFLEFKPSEIAA 362
Query: 245 AVAISLKELPTQEVD--KAITDFF-IVDKERVLKCVELIRDLSLIKV--GGNNFAS---F 296
AVA + TQ +D K+I+ V+K R+LKCV ++++SL V G ++ AS
Sbjct: 363 AVATCVVG-ETQAIDSSKSISTLIQYVEKGRLLKCVGKVQEMSLNSVFTGKDSSASSVPS 539
Query: 297 VPQSPIGVLDAGCMSFKSDELTNG 320
VPQSP+GVLD C S+KSD+ G
Sbjct: 540 VPQSPMGVLDTLCFSYKSDDTNAG 611
Score = 26.2 bits (56), Expect(2) = 4e-51
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +1
Query: 320 GSCPNSSHSSPNAKRMKFD 338
GSC +S SSP+AKR K +
Sbjct: 619 GSCSSSHSSSPDAKRRKLN 675
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 180 bits (457), Expect = 6e-46
Identities = 119/332 (35%), Positives = 181/332 (53%), Gaps = 7/332 (2%)
Frame = +2
Query: 7 SAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSD---SFLCFVAQSEEIVK 63
+++ LLC E++S +V+ D S +LD+ S S F + EE +
Sbjct: 128 TSDCELLCGEDSS----EVLTGDLPECSS------DLDSSSSSQLPSSSLFAEEEEESIA 277
Query: 64 VMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRF 123
V +E E +P DY+ R + L+ S R EA+ WI K H YYGF PL+ LSVNY+DRF
Sbjct: 278 VFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRF 457
Query: 124 LSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMI 183
L P W +QLL+VAC SLAAKMEE VP +D Q+ K++FQ +TI RMEL++
Sbjct: 458 LDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLV 637
Query: 184 LSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEIA 243
L+ L W++R++TP SF+ +F K+ +I+R+ ++IL+ I+ FL +R S IA
Sbjct: 638 LTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIA 817
Query: 244 AAVAIS-LKELPTQEV---DKAITDFFIVDKERVLKCVELIRDLSLIKVGGNNFASFVPQ 299
AA +S E+P + A + + KE+++ C ELI+++ V NN +
Sbjct: 818 AAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEI----VSSNNQRNAPKV 985
Query: 300 SPIGVLDAGCMSFKSDELTNGSCPNSSHSSPN 331
P L + + +++ S SS SSP+
Sbjct: 986 LP--QLRVTARTRRWSTVSSSSSSPSSSSSPS 1075
>TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersicon
esculentum}, partial (39%)
Length = 692
Score = 92.0 bits (227), Expect(2) = 9e-30
Identities = 53/133 (39%), Positives = 80/133 (59%), Gaps = 5/133 (3%)
Frame = +2
Query: 37 DHTNVNLDNVGSDSFLCFVAQSE-----EIVKVMVEKEKDHLPREDYLIRLRGGDLDLSV 91
++ NV+L+N ++ C + +++ E +K ++ KE+ + I L+ + +
Sbjct: 29 EYDNVSLNNTTINTTTCSLLETDMFWEDEELKSLLNKEQQN----PLYIFLQTNPVLETA 196
Query: 92 RREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKM 151
RRE+++WI K +A+Y F L+ L+VNYLDRFL F+F W QL AVAC SLAAKM
Sbjct: 197 RRESIEWILKVNAHYSFSALTSVLAVNYLDRFLFSFRFQNEKPWMTQLAAVACLSLAAKM 376
Query: 152 EEVKVPQSVDLQV 164
EE VP +DLQV
Sbjct: 377 EETHVPLLLDLQV 415
Score = 55.8 bits (133), Expect(2) = 9e-30
Identities = 23/49 (46%), Positives = 37/49 (74%)
Frame = +1
Query: 164 VGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKY 212
V E +++F+AKTI++ME++ILS+LGWKM TP SFID + ++ + +
Sbjct: 487 VEESRYLFEAKTIKKMEILILSTLGWKMNPATPLSFIDXIIRRLGLKDH 633
>BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}, partial
(50%)
Length = 663
Score = 123 bits (308), Expect = 1e-28
Identities = 81/224 (36%), Positives = 131/224 (58%), Gaps = 6/224 (2%)
Frame = +2
Query: 118 NYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQ 177
NY+DRFL+ + P+ W +QLL+VAC SLAAKMEE VP +DLQV K++F+ TI+
Sbjct: 2 NYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETLVPSLLDLQVEGVKYMFEPITIR 181
Query: 178 RMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEF 237
RMEL++LS L W++R++TP SF+ +F K+ LI+R+ Q+IL+ I+ L +
Sbjct: 182 RMELLVLSVLDWRLRSVTPFSFLSFFACKLDSTSTFTGFLISRATQIILSKIQEASILAY 361
Query: 238 RSSEI-AAAVAISLKELPT---QEVDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNNF 293
S I AAA+ + E+P E + A + + KE+++ C +L+++L +I
Sbjct: 362 WPSCIAAAAILYAANEIPNWSLVEPEHAESWCEGLRKEKIIGCYQLMQEL-VIDNNQRKP 538
Query: 294 ASFVPQSPIGV--LDAGCMSFKSDELTNGSCPNSSHSSPNAKRM 335
+PQ + + L C+S ++ S P+SS SS +++
Sbjct: 539 PKVLPQMRVTIQPLMRSCVS------SSSSSPSSSSSSYKRRKL 652
>TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhinum majus},
partial (36%)
Length = 1000
Score = 94.7 bits (234), Expect = 5e-20
Identities = 53/155 (34%), Positives = 82/155 (52%)
Frame = +3
Query: 9 ESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMVEK 68
+S+LLC E S+ ++ DS D + + +E + ++E
Sbjct: 186 DSDLLCGEETSS----ILSSDSPTESFSDGES-------------YPPPEDEFIAGLIED 314
Query: 69 EKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQ 128
+ + DY ++++ D R E++ WI K YYGF P++ L+VNY+DRFL+ +
Sbjct: 315 QGKFVIGFDYFVKMKSSSFDSDARDESIRWILKVQGYYGFQPVTAYLAVNYMDRFLNSRR 494
Query: 129 FPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQ 163
P+ W +QLL+VAC SLAAKMEE VP +DLQ
Sbjct: 495 LPQTNGWPLQLLSVACLSLAAKMEETLVPSLLDLQ 599
>TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalfa, partial
(51%)
Length = 1011
Score = 75.1 bits (183), Expect = 4e-14
Identities = 55/184 (29%), Positives = 91/184 (48%), Gaps = 8/184 (4%)
Frame = +1
Query: 170 VFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNII 229
VF+AKTIQRMEL+ILS+L WKM +T SF+D+ + ++ + + R L+L+++
Sbjct: 1 VFEAKTIQRMELLILSTLKWKMHPVTTHSFLDHIIRRLGLKTNLHWEFLRRCENLLLSVL 180
Query: 230 KGIDFLEFRSSEIAAAVAI----SLKELPTQEVD--KAITDFFIVDKERVLKCVELIRDL 283
F+ S +A A + +++ +VD + + + KE+V +C I L
Sbjct: 181 LDSRFVGCVPSVLATATMLHVIDQIEQSDDNDVDYKNQLLNVLKISKEKVDECYNAILHL 360
Query: 284 SLIKVGGNNFASF--VPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNAKRMKFDGPS 341
+ G N + +P SP GV+DA S S++ + S S P K+ K G +
Sbjct: 361 TNTNNYGYNKRKYEEIPGSPSGVIDAVFSSDGSNDSWTVGASSYSTSEPVFKKTKNQGQN 540
Query: 342 SGTS 345
S
Sbjct: 541 MNLS 552
>TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medicago sativa},
partial (41%)
Length = 689
Score = 66.2 bits (160), Expect = 2e-11
Identities = 42/115 (36%), Positives = 60/115 (51%), Gaps = 9/115 (7%)
Frame = +1
Query: 39 TNVNLDNVGSDSFLCFVA------QSEEIVKVMVEKEKDHLPREDY---LIRLRGGDLDL 89
T N D + S S L + +E + + KEK + +E Y L + D
Sbjct: 346 TTTNNDILDSTSLLPLLLLEQNLFNKDEELNTLFSKEK--IQQETYYDDLKNVMNFDSLT 519
Query: 90 SVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVAC 144
RREA++W+ K +A+YGF L+ L+VNYLDRFL F F + W +QL+AV C
Sbjct: 520 QPRREAVEWMLKVNAHYGFSALTATLAVNYLDRFLLSFHFQKEKPWMIQLVAVTC 684
>TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersicon
esculentum}, partial (30%)
Length = 637
Score = 65.1 bits (157), Expect = 4e-11
Identities = 52/169 (30%), Positives = 88/169 (51%), Gaps = 11/169 (6%)
Frame = +3
Query: 171 FQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKY-PDKSLIARSVQLILNII 229
F+AKTI++ME++ILS+LGWKM TP SFID+ + ++ + + + R ++L++I
Sbjct: 9 FEAKTIKKMEILILSTLGWKMNPATPLSFIDFIIRRLGLKDHLICWEFLKRCEGVLLSVI 188
Query: 230 KG-IDFLEFRSSEIAAAVAI----SLKELPTQEVDKAITDFFIVDKERVLKCVELIRDLS 284
+ F+ + S +A A + S++ E + ++K++V +C +L+ L
Sbjct: 189 RSDSKFMSYLPSVLATATMVHVFNSVEPSLGDEYQTQLLGILGINKDKVDECGKLLLKLW 368
Query: 285 LIKVGGN-----NFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHS 328
GN F S +P SP GV++ MSF SC NS+ S
Sbjct: 369 SGYEEGNECNKRKFGS-IPSSPKGVME---MSF--------SCDNSNDS 479
>BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%)
Length = 519
Score = 60.5 bits (145), Expect = 9e-10
Identities = 37/114 (32%), Positives = 58/114 (50%)
Frame = +3
Query: 38 HTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALD 97
HT++ D + + S+++ + + E DH+P +Y L+ + D SVR + +
Sbjct: 123 HTSMEFDLENPLEYFHDLPNSQDVSSLFLI-ESDHIPPLNYFQNLKSNEFDASVRTDFIS 299
Query: 98 WIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKM 151
I + F P L++NYLDRFL+ + W +LLAV CFSLA KM
Sbjct: 300 LISQLSC--NFDPFVTYLAINYLDRFLANQGILQPKPWANKLLAVTCFSLAVKM 455
>AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (33%)
Length = 648
Score = 57.4 bits (137), Expect = 8e-09
Identities = 38/124 (30%), Positives = 64/124 (50%)
Frame = +3
Query: 38 HTNVNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALD 97
HT +N +V+ + ++ M E +K P YL ++ G + ++R +D
Sbjct: 285 HTKINAKRDNQHIIQPYVSDISDYLRTM-EMQKKRRPMVGYLENVQRG-ISSNMRGTLVD 458
Query: 98 WIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVP 157
W+ + Y P +L LSV+Y+DRFLS+ R +QLL V+ +A+K EE+ P
Sbjct: 459 WLVEVADEYKLLPETLHLSVSYIDRFLSIEPVSRS---KLQLLGVSSMLIASKYEEITPP 629
Query: 158 QSVD 161
++VD
Sbjct: 630 KAVD 641
>TC80999 similar to GP|10176986|dbj|BAB10218.
gene_id:MYH19.13~pir||S76678~similar to unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 707
Score = 29.6 bits (65), Expect = 1.8
Identities = 16/43 (37%), Positives = 23/43 (53%)
Frame = -2
Query: 239 SSEIAAAVAISLKELPTQEVDKAITDFFIVDKERVLKCVELIR 281
S E A IS + P + D A+T FF+V E L+C ++R
Sbjct: 652 SEEPAGICLISRRNFPQDKTDTAVT-FFLVCMETPLRCTSVMR 527
>BE204732 weakly similar to GP|8978275|db receptor protein kinase-like
{Arabidopsis thaliana}, partial (10%)
Length = 461
Score = 28.9 bits (63), Expect = 3.0
Identities = 15/48 (31%), Positives = 23/48 (47%)
Frame = -1
Query: 171 FQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLI 218
F+ + R I+S +G L C F+ +F IS + P KSL+
Sbjct: 416 FKDRNTSRCRFPIVSGIGPVKPQLISCKFVSFFKFPISAGRGPVKSLL 273
>TC81552 similar to GP|16755592|gb|AAL28022.1 small GTPase Rab2 {Nicotiana
tabacum}, complete
Length = 887
Score = 28.5 bits (62), Expect = 4.0
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = +2
Query: 158 QSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTP 196
QS+DL E + F T+Q +I+ +GWKMR P
Sbjct: 281 QSLDLTTEEQQGRF*FMTLQGERHLIIWQVGWKMRGSMP 397
>TC92830 weakly similar to PIR|A85043|A85043 probable LRR receptor-like
protein kinase [imported] - Arabidopsis thaliana,
partial (11%)
Length = 512
Score = 28.1 bits (61), Expect = 5.2
Identities = 18/56 (32%), Positives = 24/56 (42%)
Frame = -1
Query: 275 KCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSP 330
KC L+RD+ LI+ G + P + CM S+ L C S H SP
Sbjct: 509 KCGLLLRDIDLIETNGPKLSPNTPPKFAPIRVISCM---SESLH*TPCHASPHGSP 351
>TC83095 similar to GP|9293910|dbj|BAB01813.1 gene_id:MXO21.1~unknown
protein {Arabidopsis thaliana}, partial (45%)
Length = 808
Score = 27.7 bits (60), Expect = 6.8
Identities = 17/63 (26%), Positives = 35/63 (54%), Gaps = 2/63 (3%)
Frame = +1
Query: 4 SFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVG--SDSFLCFVAQSEEI 61
SF+ + +++ CS ++ +CF +D++ +S + +NL N+G + SF F A +
Sbjct: 76 SFNPSSNSIKCSASSFSCFSSTSLDETAMSIENLRSFINL-NIGKWNGSFYQFDAGGNPL 252
Query: 62 VKV 64
+V
Sbjct: 253 QRV 261
>TC93226 weakly similar to PIR|B96737|B96737 hypothetical protein F3I17.10
[imported] - Arabidopsis thaliana, partial (60%)
Length = 847
Score = 27.3 bits (59), Expect = 8.9
Identities = 23/72 (31%), Positives = 36/72 (49%), Gaps = 3/72 (4%)
Frame = -1
Query: 141 AVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRA---LTPC 197
AV C +L+ K E K+ + D + +F+++ I + ++I SS LT C
Sbjct: 517 AV*CQNLSPKGIEKKIAKLRDRIISTSAIIFRSEVI--IYIIITSSHHNHY*LS*ELTQC 344
Query: 198 SFIDYFLAKISC 209
SFI +FL I C
Sbjct: 343 SFIHHFLILIQC 308
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,898,549
Number of Sequences: 36976
Number of extensions: 149588
Number of successful extensions: 773
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 767
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 768
length of query: 346
length of database: 9,014,727
effective HSP length: 97
effective length of query: 249
effective length of database: 5,428,055
effective search space: 1351585695
effective search space used: 1351585695
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135848.10


