

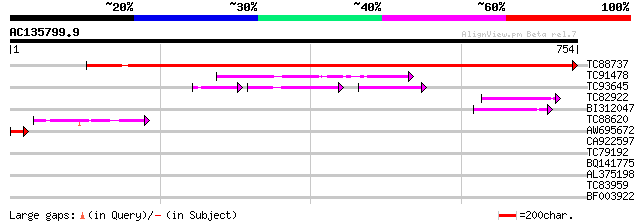
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135799.9 - phase: 0
(754 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88737 similar to PIR|A96737|A96737 hypothetical protein F3I17.... 1271 0.0
TC91478 similar to GP|15810447|gb|AAL07111.1 unknown protein {Ar... 99 6e-21
TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2 ... 46 2e-18
TC82922 weakly similar to PIR|B96526|B96526 unknown protein [imp... 56 4e-08
BI312047 weakly similar to PIR|D84604|D846 hypothetical protein ... 55 1e-07
TC88620 weakly similar to GP|15810447|gb|AAL07111.1 unknown prot... 54 3e-07
AW695672 GP|13183344|g vitellogenin A {Melanogrammus aeglefinus}... 50 2e-06
CA922597 34 0.18
TC79192 similar to GP|14517387|gb|AAK62584.1 AT4g13980/dl3030c {... 33 0.53
BQ141775 31 1.5
AL375198 homologue to PIR|T11622|T116 extensin class 1 precursor... 30 4.5
TC83959 similar to GP|21593908|gb|AAM65873.1 pore protein homolo... 29 5.9
BF003922 similar to GP|5081557|gb|A PHAP2B protein {Petunia x hy... 28 10.0
>TC88737 similar to PIR|A96737|A96737 hypothetical protein F3I17.11
[imported] - Arabidopsis thaliana, partial (62%)
Length = 2115
Score = 1271 bits (3290), Expect = 0.0
Identities = 645/652 (98%), Positives = 645/652 (98%)
Frame = +1
Query: 103 FLSRDNSDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQVAWVTLQSKRVTE 162
FLSRDNSDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQ SKRVTE
Sbjct: 1 FLSRDNSDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQ-------SKRVTE 159
Query: 163 EAFVRIAPAVSGVVDRPTVHNLFKVLAGDKDGISMSTWLAYINEFVKVRRENRSYQIPEF 222
EAFVRIAPAVSGVVDRPTVHNLFKVLAGDKDGISMSTWLAYINEFVKVRRENRSYQIPEF
Sbjct: 160 EAFVRIAPAVSGVVDRPTVHNLFKVLAGDKDGISMSTWLAYINEFVKVRRENRSYQIPEF 339
Query: 223 PQIDEEKILCIGSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYD 282
PQIDEEKILCIGSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYD
Sbjct: 340 PQIDEEKILCIGSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYD 519
Query: 283 GLRVEKAKVGPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHK 342
GLRVEKAKVGPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHK
Sbjct: 520 GLRVEKAKVGPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHK 699
Query: 343 FTHEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTKLVQFSYLQNA 402
FTHEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTKLVQFSYLQNA
Sbjct: 700 FTHEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTKLVQFSYLQNA 879
Query: 403 PNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWMKS 462
PNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWMKS
Sbjct: 880 PNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWMKS 1059
Query: 463 PSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATL 522
PSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATL
Sbjct: 1060PSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATL 1239
Query: 523 KGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVML 582
KGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVML
Sbjct: 1240KGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVML 1419
Query: 583 MITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNVS 642
MITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNVS
Sbjct: 1420MITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNVS 1599
Query: 643 LLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMVER 702
LLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMVER
Sbjct: 1600LLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMVER 1779
Query: 703 LTKLLRERWHAVPAAPVAVLPFENEESKSEVSLKELENKSKPPGNQSKRKSG 754
LTKLLRERWHAVPAAPVAVLPFENEESKSEVSLKELENKSKPPGNQSKRKSG
Sbjct: 1780LTKLLRERWHAVPAAPVAVLPFENEESKSEVSLKELENKSKPPGNQSKRKSG 1935
>TC91478 similar to GP|15810447|gb|AAL07111.1 unknown protein {Arabidopsis
thaliana}, partial (27%)
Length = 736
Score = 99.0 bits (245), Expect = 6e-21
Identities = 82/264 (31%), Positives = 127/264 (48%), Gaps = 2/264 (0%)
Frame = +3
Query: 276 RLDLTYDGLRVEKAKV-GPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALI 334
R DL D +V K ++ GP G+ LFD AV SS S S VLEF +L G RRD W A+I
Sbjct: 36 RYDLADDLNQVVKPELTGPWGTRLFDKAVFYSSTSLSEPAVLEFPELKGHARRDYWLAII 215
Query: 335 SEVIALHKFTHEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPT-KL 393
E++ +HK+ ++G + A A+ GI RLQA+Q + L P L
Sbjct: 216 REILCVHKYISKFGIKGV---------ARDEALWKAVLGILRLQAIQDISSLGPVPNDSL 368
Query: 394 VQFSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGS 453
+ F+ P GD++L++L VN ++ + + H +++P + + S V D +
Sbjct: 369 LMFNLCDQLPGGDLILETL-VN------MSNSSESNHGHDSKPESGMYSIS--VLDTVSN 521
Query: 454 VYLRKWMKSPSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKT 513
+ G TS N+S + + V + +L ER K SK + V
Sbjct: 522 L-----------GFVFGTS--SNSSVESRIAVGEITVGETTLLERVVKESKNNYKKVVSA 662
Query: 514 QATIDAATLKGIPSNIDLFKELIF 537
QAT D + GI +N+ + KEL++
Sbjct: 663 QATSDGVKVDGIDTNLAVMKELLY 734
>TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2
{Arabidopsis thaliana}, partial (26%)
Length = 994
Score = 45.8 bits (107), Expect(3) = 2e-18
Identities = 28/92 (30%), Positives = 47/92 (50%), Gaps = 2/92 (2%)
Frame = +1
Query: 465 WGSSTSTSFWKNT--STKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATL 522
W SS S K S K +L + V +++ + A K S + E QAT+D +
Sbjct: 577 WHSSVLDSNQKRRLISMKKQLLVCDIRVGEINPLDMAVKQSLLDTGKAEAAQATVDQVKV 756
Query: 523 KGIPSNIDLFKELIFPITLTAKNFEKLRHWEE 554
+GI +N+ + KEL+FP+ +A + L W++
Sbjct: 757 EGIDTNVAVMKELLFPVIESANRLQLLASWKD 852
Score = 43.1 bits (100), Expect(3) = 2e-18
Identities = 30/129 (23%), Positives = 57/129 (43%), Gaps = 1/129 (0%)
Frame = +3
Query: 317 EFIDLGGDMRRDVWHALISEVIALHKFTHEYGPDEYGPNVFEARKGKQRATSSAINGIAR 376
EF + ++RRD W + E++ H+F ++G + K + A GI R
Sbjct: 213 EFPEFKANLRRDYWLDISLEILRAHRFVRKFGLKD---------TQKLEVLARANLGIFR 365
Query: 377 LQALQHLRKLLDDPTK-LVQFSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENR 435
+AL+ K K L+ F+ + P GD +LQ+L+ + S +G E +
Sbjct: 366 YRALKEAFKFFSSNYKTLLTFNLAETLPRGDRILQTLSNSLTNSTAASGKRDISANVERK 545
Query: 436 PSNEIADSS 444
+ ++ ++
Sbjct: 546 KQSAVSPAA 572
Score = 42.0 bits (97), Expect(3) = 2e-18
Identities = 27/67 (40%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Frame = +2
Query: 244 WENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGLRVEKAKV-GPLGSSLFDSA 302
W MAW A+YFE G+ +++A+R DL D +V K ++ GPLG+ LFD A
Sbjct: 2 WNCRMAWA--FDPDQYALYFESLGVGVHEKAIRYDLGADMKQVIKPELTGPLGARLFDKA 175
Query: 303 VSISSGS 309
V S S
Sbjct: 176 VMYKSTS 196
>TC82922 weakly similar to PIR|B96526|B96526 unknown protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 648
Score = 56.2 bits (134), Expect = 4e-08
Identities = 35/105 (33%), Positives = 54/105 (51%)
Frame = +1
Query: 628 AMRDVENMTQKVNVSLLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFD 687
A+ E N+ LLK R + LS PQ T ++A +L+ +L +P KYI+ + +
Sbjct: 1 AVSQAEQFILDGNLFLLKFRGLWLSFFPQATDKMAFGLLSAGLVLAFLPTKYIVLLVFLN 180
Query: 688 MFTRELEFRREMVERLTKLLRERWHAVPAAPVAVLPFENEESKSE 732
FT R+ ER + RE W ++PAAPV L + E+ K +
Sbjct: 181 TFTMFSPPRKASTERWERRFREWWFSIPAAPVK-LERDKEDKKKK 312
>BI312047 weakly similar to PIR|D84604|D846 hypothetical protein At2g21720
[imported] - Arabidopsis thaliana, partial (12%)
Length = 640
Score = 55.1 bits (131), Expect = 1e-07
Identities = 32/106 (30%), Positives = 56/106 (52%)
Frame = +3
Query: 617 NTIQKIIAVKDAMRDVENMTQKVNVSLLKIRSILLSGNPQITTEVAVLMLTWATILFIVP 676
+T+ I++ + + V M Q N+++LKI SIL+S + V V M A +L ++P
Sbjct: 66 STMDSIVSAQHGLYTVHEMMQIANIAMLKIWSILISKADKHANAVMVAMGGLAFLLAVIP 245
Query: 677 FKYILSFLLFDMFTRELEFRREMVERLTKLLRERWHAVPAAPVAVL 722
FK+ L L+ FT L+ + + L+E W ++P P+ V+
Sbjct: 246 FKFFLMGLIVQSFTMTLKTGKSSGTG-NRRLKEWWDSIPIVPIRVV 380
>TC88620 weakly similar to GP|15810447|gb|AAL07111.1 unknown protein
{Arabidopsis thaliana}, partial (25%)
Length = 785
Score = 53.5 bits (127), Expect = 3e-07
Identities = 47/157 (29%), Positives = 72/157 (44%), Gaps = 3/157 (1%)
Frame = +2
Query: 32 FQVMEDILMVEKTIDRKMPYGNLSLAAVICIEQFSRMSGLTGKKMKNIFETLVPETVYND 91
F+ ME+ K R+ LS A + + + +++ + ++ F +++ N
Sbjct: 296 FEEMENSPSAGKNFVRE-----LSPVANLVVRRCAKILKTSSSDLQEGFNLEASDSLKNP 460
Query: 92 ---ARNLVEYCCFRFLSRDNSDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASL 148
ARNL+EYCCF+ LS + + L D F+RL + MLAWE P A+
Sbjct: 461 SRYARNLLEYCCFKALSL-KAQISGHLLDKTFRRLTYDMMLAWEVP----------AATS 607
Query: 149 QVAWVTLQSKRVTEEAFVRIAPAVSGVVDRPTVHNLF 185
Q + V EAF RIAPAV + + NLF
Sbjct: 608 QPLTNVDEEVSVGLEAFCRIAPAVPIIANVIICENLF 718
>AW695672 GP|13183344|g vitellogenin A {Melanogrammus aeglefinus}, partial
(0%)
Length = 371
Score = 50.4 bits (119), Expect = 2e-06
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = +1
Query: 1 MSRTQNFLNEVTSPLAKTAQSRKP 24
MSRTQNFLNEVTSPLAKTAQSRKP
Sbjct: 298 MSRTQNFLNEVTSPLAKTAQSRKP 369
>CA922597
Length = 750
Score = 34.3 bits (77), Expect = 0.18
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = -2
Query: 687 DMFTRELEFRREMVERLTKLLRERWHAVPAAPVAVL 722
+ +TRE+ RRE +R + RE W +PAAPV ++
Sbjct: 749 ECYTREMPCRRESSKRWIRRFREWWIRIPAAPVELV 642
>TC79192 similar to GP|14517387|gb|AAK62584.1 AT4g13980/dl3030c {Arabidopsis
thaliana}, partial (57%)
Length = 1686
Score = 32.7 bits (73), Expect = 0.53
Identities = 26/102 (25%), Positives = 45/102 (43%), Gaps = 2/102 (1%)
Frame = -2
Query: 53 NLSLAAVICIEQFSRMSGLTGKKMKNIFETLVPETVYNDARNLVEYCCFRFLSRDNSDVH 112
+LS + + +FS+ + T + + TL PE + AR + FRF SD
Sbjct: 1373 SLSEQPGLSVRKFSQNTSFTPCRAAALTWTL-PEASFGGARLIASLATFRFEETPISDSL 1197
Query: 113 PSLQDPAFQRLIFITMLAWEN--PYTYVLSSNAEKASLQVAW 152
PS +L W + Y ++L+SN + A ++ +W
Sbjct: 1196 PSALTLDSAKLPIS*QFIWGDLAEYEFLLNSNVQDARVKFSW 1071
>BQ141775
Length = 727
Score = 31.2 bits (69), Expect = 1.5
Identities = 12/38 (31%), Positives = 22/38 (57%)
Frame = -1
Query: 456 LRKWMKSPSWGSSTSTSFWKNTSTKGLVLSKNHVVADL 493
L KW +P+W S + ++N + V S +H++A+L
Sbjct: 646 LSKWCNNPTWVSRVKSCIYENLFHQASVNSYHHIIAEL 533
>AL375198 homologue to PIR|T11622|T116 extensin class 1 precursor - cowpea,
partial (8%)
Length = 684
Score = 29.6 bits (65), Expect = 4.5
Identities = 14/60 (23%), Positives = 25/60 (41%)
Frame = +1
Query: 79 IFETLVPETVYNDARNLVEYCCFRFLSRDNSDVHPSLQDPAFQRLIFITMLAWENPYTYV 138
+FET + +Y R+ YC F S+ + H + + P ++ + P YV
Sbjct: 445 VFETCLLSRLYKSNRDTARYCPFSRRSQQTTHTHKTTRPPHAASVLXVLTPRLSGPLKYV 624
>TC83959 similar to GP|21593908|gb|AAM65873.1 pore protein homolog
{Arabidopsis thaliana}, partial (73%)
Length = 725
Score = 29.3 bits (64), Expect = 5.9
Identities = 25/113 (22%), Positives = 50/113 (44%), Gaps = 5/113 (4%)
Frame = -1
Query: 578 FPVMLMITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQ 637
FPV++ + + +L + L+ G+ G PP +T+ +AVK A R+ T
Sbjct: 356 FPVLITMASRDLLLVSPLRPAGKRRLLVGETSCGITPPLSTL*PSMAVK*ASRETA-*TA 180
Query: 638 KVNVSLLKIRSILL-----SGNPQITTEVAVLMLTWATILFIVPFKYILSFLL 685
+ +L K+ ++ L N +++ + ++ FK+I +LL
Sbjct: 179 PIPAALTKLSAMRLRRGWPRSNRPFLSKLRISSSRALVSTSLLVFKFIFDYLL 21
>BF003922 similar to GP|5081557|gb|A PHAP2B protein {Petunia x hybrida},
partial (11%)
Length = 615
Score = 28.5 bits (62), Expect = 10.0
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = -3
Query: 314 WVLEFIDLGGDMRRDVWHALISE 336
W +EFIDLGG+MR D A + E
Sbjct: 556 WGIEFIDLGGNMRVDGMGAELME 488
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,913,357
Number of Sequences: 36976
Number of extensions: 266125
Number of successful extensions: 1487
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1474
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1483
length of query: 754
length of database: 9,014,727
effective HSP length: 103
effective length of query: 651
effective length of database: 5,206,199
effective search space: 3389235549
effective search space used: 3389235549
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135799.9


