

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135798.2 + phase: 0
(261 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
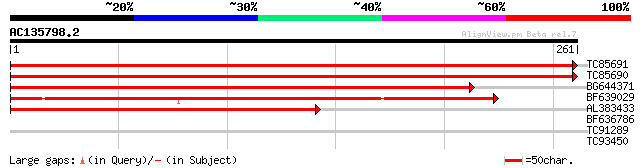
Sequences producing significant alignments: (bits) Value
TC85691 similar to GP|6714564|dbj|BAA89498.1 cyc07 {Daucus carot... 520 e-148
TC85690 similar to SP|P33444|RS3A_CATRO 40S ribosomal protein S3... 487 e-138
BG644371 homologue to GP|6714564|dbj cyc07 {Daucus carota}, part... 384 e-107
BF639029 similar to PIR|S36622|S36 ribosomal protein S3a - turni... 292 1e-79
AL383433 similar to PIR|S49366|S49 ribosomal protein S0.e.B cyt... 220 4e-58
BF636786 similar to PIR|T49132|T49 hypothetical protein F26G5.11... 30 1.2
TC91289 similar to GP|10177361|dbj|BAB10652. cell cycle control ... 29 2.1
TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associa... 27 7.9
>TC85691 similar to GP|6714564|dbj|BAA89498.1 cyc07 {Daucus carota},
complete
Length = 1159
Score = 520 bits (1339), Expect = e-148
Identities = 261/261 (100%), Positives = 261/261 (100%)
Frame = +3
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG
Sbjct: 57 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 236
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL
Sbjct: 237 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 416
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD
Sbjct: 417 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 596
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG
Sbjct: 597 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 776
Query: 241 TKVERPADETMVEGTPEIVGA 261
TKVERPADETMVEGTPEIVGA
Sbjct: 777 TKVERPADETMVEGTPEIVGA 839
>TC85690 similar to SP|P33444|RS3A_CATRO 40S ribosomal protein S3A (CYC07
protein). [Rosy periwinkle Madagascar periwinkle],
partial (97%)
Length = 1147
Score = 487 bits (1253), Expect = e-138
Identities = 244/261 (93%), Positives = 253/261 (96%)
Frame = +3
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKAADPF+KKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIAS+G
Sbjct: 69 MAVGKNKRISKGKKGGKKKAADPFSKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASDG 248
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDE++AFRKIRLRAEDVQGKNLLTNFWGM+FTTDKLRSLVRKWQTL
Sbjct: 249 LKHRVFEVSLADLQGDEEHAFRKIRLRAEDVQGKNLLTNFWGMNFTTDKLRSLVRKWQTL 428
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNYTLRMFCIGFTKRR+NQVKRTCYAQSSQIRQIRRKM EIMINQA+SCD
Sbjct: 429 IEAHVDVKTTDNYTLRMFCIGFTKRRANQVKRTCYAQSSQIRQIRRKMVEIMINQASSCD 608
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK LV KFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY+EDVG
Sbjct: 609 LKGLVHKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYNEDVG 788
Query: 241 TKVERPADETMVEGTPEIVGA 261
TKVERPADE + E EIVGA
Sbjct: 789 TKVERPADEMVTEEPTEIVGA 851
>BG644371 homologue to GP|6714564|dbj cyc07 {Daucus carota}, partial (81%)
Length = 729
Score = 384 bits (985), Expect = e-107
Identities = 191/214 (89%), Positives = 204/214 (95%)
Frame = +3
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKGGKKKAADP+AKKDWYDIKAPSVF++KNVGKTLV+RTQGTKIASEG
Sbjct: 81 MAVGKNKRISKGKKGGKKKAADPYAKKDWYDIKAPSVFEIKNVGKTLVTRTQGTKIASEG 260
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQ DED AFRKIRLRAEDVQG+N+LTNF GMDFTTDKLRSLVRKWQTL
Sbjct: 261 LKHRVFEVSLADLQKDEDQAFRKIRLRAEDVQGRNVLTNFHGMDFTTDKLRSLVRKWQTL 440
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD+YTLRMFCI FTK+R NQ KRTCYAQSSQIRQIRRKM EIM NQA+SCD
Sbjct: 441 IEAHVDVKTTDSYTLRMFCIAFTKKRPNQQKRTCYAQSSQIRQIRRKMVEIMRNQASSCD 620
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRK 214
LK+LV KFIPE IG+EIEKATSSI+PLQNV+IRK
Sbjct: 621 LKELVAKFIPESIGREIEKATSSIFPLQNVYIRK 722
>BF639029 similar to PIR|S36622|S36 ribosomal protein S3a - turnip, partial
(74%)
Length = 691
Score = 292 bits (747), Expect = 1e-79
Identities = 148/226 (65%), Positives = 179/226 (78%), Gaps = 1/226 (0%)
Frame = +1
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNK++ K +KG +K DPFAKK+WYDIKAP+VF ++ VGKT+V++TQGTK + +
Sbjct: 19 MAVGKNKKLGKKRKG-TRKITDPFAKKEWYDIKAPAVFPIRQVGKTVVTKTQGTKNSRDS 195
Query: 61 LKHRVFEVSLADLQGD-EDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQT 119
L RVFE SL DL+ ED AFRK RL+ EDVQG NLLT F+GMD TTDKLRSLVRKW +
Sbjct: 196 LMGRVFEASLGDLKPHGEDEAFRKFRLKVEDVQGTNLLTQFYGMDMTTDKLRSLVRKWHS 375
Query: 120 LIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSC 179
LIE HVDVKTTD YTLR+F IGFTKRR NQ ++T YAQ++Q+R IR+K I + ++C
Sbjct: 376 LIETHVDVKTTDGYTLRLFVIGFTKRRPNQNRKTSYAQTAQVRAIRKKFIHI-AQRESNC 552
Query: 180 DLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDL 225
DL +LV KFIP +IGKEIEKAT IYPLQNVFIRKVK L+AP D+
Sbjct: 553 DLNELVNKFIPXIIGKEIEKATQGIYPLQNVFIRKVKTLRAPXVDV 690
>AL383433 similar to PIR|S49366|S49 ribosomal protein S0.e.B cytosolic -
yeast (Candida albicans), partial (55%)
Length = 471
Score = 220 bits (561), Expect = 4e-58
Identities = 106/143 (74%), Positives = 123/143 (85%)
Frame = +3
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKR+SKGKKG KKK DPF +KDWYD+KAPS+F+V+N+GKTLV+RTQG K A+
Sbjct: 42 MAVGKNKRLSKGKKGLKKKIVDPFTRKDWYDVKAPSIFEVRNIGKTLVNRTQGLKNANXA 221
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LK RVFEVSLADL DE+ AFRKI+LR ++VQGKNLLTNF GMDFT+ KLRSLV+KWQ+L
Sbjct: 222 LKGRVFEVSLADLNKDEEQAFRKIKLRVDEVQGKNLLTNFHGMDFTSXKLRSLVKKWQSL 401
Query: 121 IEAHVDVKTTDNYTLRMFCIGFT 143
IEAHVDV TTD Y LR+F I FT
Sbjct: 402 IEAHVDVXTTDGYLLRLFAIAFT 470
>BF636786 similar to PIR|T49132|T49 hypothetical protein F26G5.110 -
Arabidopsis thaliana, partial (24%)
Length = 567
Score = 29.6 bits (65), Expect = 1.2
Identities = 16/48 (33%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Frame = +3
Query: 128 KTTDNYTLRMFCIGFTKRRSNQVKRTCY--AQSSQIRQIRRKMREIMI 173
+T N TLR+FC T R + Q K T Y + S++ + R +M+
Sbjct: 15 RTHKNTTLRLFCFPLTLRNTTQHKHTLYI*SNLSKVPNLLRNCFYLMV 158
>TC91289 similar to GP|10177361|dbj|BAB10652. cell cycle control crn
(crooked neck) protein-like {Arabidopsis thaliana},
partial (20%)
Length = 698
Score = 28.9 bits (63), Expect = 2.1
Identities = 22/63 (34%), Positives = 29/63 (45%), Gaps = 2/63 (3%)
Frame = +2
Query: 62 KHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRK--WQT 119
K FE D +G ED K R + ED KNLL +G+ K S +RK W+
Sbjct: 212 KFVAFEKQYGDREGIEDAIVGKRRFQYEDEVRKNLLNYDYGLII*DGKRVSGIRKELWRF 391
Query: 120 LIE 122
+ E
Sbjct: 392 MRE 400
>TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associated
protein-like {Arabidopsis thaliana}, partial (22%)
Length = 806
Score = 26.9 bits (58), Expect = 7.9
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = +3
Query: 208 QNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVGTKVERPADETMVEG 254
Q ++R +++L +F L KL+ + + +KVE ADE + G
Sbjct: 180 QYFYLRWLRVLFGREFSLDKLLVIWDEIFASDNSKVESSADENIDYG 320
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,124,302
Number of Sequences: 36976
Number of extensions: 65127
Number of successful extensions: 331
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 328
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 328
length of query: 261
length of database: 9,014,727
effective HSP length: 94
effective length of query: 167
effective length of database: 5,538,983
effective search space: 925010161
effective search space used: 925010161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135798.2


