

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135796.9 - phase: 0 /pseudo
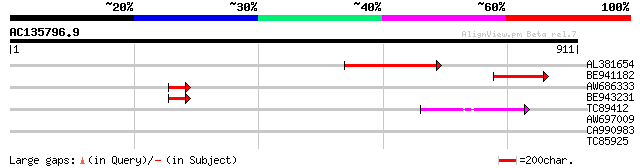
(911 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AL381654 similar to GP|9759599|db DNA-directed RNA polymerase su... 302 3e-82
BE941182 similar to GP|9759599|dbj DNA-directed RNA polymerase s... 172 5e-43
AW686333 similar to GP|9759599|dbj DNA-directed RNA polymerase s... 68 2e-11
BE943231 similar to GP|9759599|dbj DNA-directed RNA polymerase s... 68 2e-11
TC89412 similar to GP|9293951|dbj|BAB01854.1 DNA-directed RNA po... 54 3e-07
AW697009 similar to PIR|D96805|D96 probable acyl-CoA synthetase ... 34 0.29
CA990983 similar to GP|22202663|dbj putative elicitor response p... 29 7.1
TC85925 weakly similar to GP|20259099|gb|AAM14265.1 putative ubi... 29 9.3
>AL381654 similar to GP|9759599|db DNA-directed RNA polymerase subunit
{Arabidopsis thaliana}, partial (9%)
Length = 473
Score = 302 bits (774), Expect = 3e-82
Identities = 157/157 (100%), Positives = 157/157 (100%)
Frame = +1
Query: 538 S*SRLWPLYSYEKVCRCPTKVFKWHRRSNRKAR*SFKQYADT***WDCFTWRNY*AN*HL 597
S*SRLWPLYSYEKVCRCPTKVFKWHRRSNRKAR*SFKQYADT***WDCFTWRNY*AN*HL
Sbjct: 1 S*SRLWPLYSYEKVCRCPTKVFKWHRRSNRKAR*SFKQYADT***WDCFTWRNY*AN*HL 180
Query: 598 YQQEITN*NTE*SVVN*LA**CISV*LFNIQ*MSWR*SG**SSSLQ**G*QHVYQISNPS 657
YQQEITN*NTE*SVVN*LA**CISV*LFNIQ*MSWR*SG**SSSLQ**G*QHVYQISNPS
Sbjct: 181 YQQEITN*NTE*SVVN*LA**CISV*LFNIQ*MSWR*SG**SSSLQ**G*QHVYQISNPS 360
Query: 658 HSEARAWG*IQ*QTWAERCLRNNCPAGGLPIF*KRHL 694
HSEARAWG*IQ*QTWAERCLRNNCPAGGLPIF*KRHL
Sbjct: 361 HSEARAWG*IQ*QTWAERCLRNNCPAGGLPIF*KRHL 471
>BE941182 similar to GP|9759599|dbj DNA-directed RNA polymerase subunit
{Arabidopsis thaliana}, partial (7%)
Length = 265
Score = 172 bits (436), Expect = 5e-43
Identities = 88/88 (100%), Positives = 88/88 (100%)
Frame = +1
Query: 778 FYGANILPEIKTHGAG*DACSW*WP*SPDN*TANRRES*KWRSTSRRNGT*LLNCIWC*Y 837
FYGANILPEIKTHGAG*DACSW*WP*SPDN*TANRRES*KWRSTSRRNGT*LLNCIWC*Y
Sbjct: 1 FYGANILPEIKTHGAG*DACSW*WP*SPDN*TANRRES*KWRSTSRRNGT*LLNCIWC*Y 180
Query: 838 VDF*ATHVI**SL*SSGL*SLWIVGILQ 865
VDF*ATHVI**SL*SSGL*SLWIVGILQ
Sbjct: 181 VDF*ATHVI**SL*SSGL*SLWIVGILQ 264
>AW686333 similar to GP|9759599|dbj DNA-directed RNA polymerase subunit
{Arabidopsis thaliana}, partial (4%)
Length = 596
Score = 67.8 bits (164), Expect = 2e-11
Identities = 34/36 (94%), Positives = 34/36 (94%)
Frame = +2
Query: 255 ALSL*YS*R*ILWTSKEPGFDDSCHY**RGGPPDFS 290
ALSL*YS*R*ILWTSKEPGFDDSCH **RGG PDFS
Sbjct: 287 ALSL*YS*R*ILWTSKEPGFDDSCHC**RGGTPDFS 394
>BE943231 similar to GP|9759599|dbj DNA-directed RNA polymerase subunit
{Arabidopsis thaliana}, partial (4%)
Length = 434
Score = 67.8 bits (164), Expect = 2e-11
Identities = 34/36 (94%), Positives = 34/36 (94%)
Frame = +3
Query: 255 ALSL*YS*R*ILWTSKEPGFDDSCHY**RGGPPDFS 290
ALSL*YS*R*ILWTSKEPGFDDSCH **RGG PDFS
Sbjct: 174 ALSL*YS*R*ILWTSKEPGFDDSCHC**RGGTPDFS 281
>TC89412 similar to GP|9293951|dbj|BAB01854.1 DNA-directed RNA polymerase
subunit B {Arabidopsis thaliana}, partial (33%)
Length = 1251
Score = 53.9 bits (128), Expect = 3e-07
Identities = 48/176 (27%), Positives = 71/176 (40%)
Frame = +3
Query: 660 EARAWG*IQ*QTWAERCLRNNCPAGGLPIF*KRHLSRSDYESSWVSKSNDSWKDDRASWR 719
E +W *+ W E C + + + RH SR Y+S+ +S + +SW R +
Sbjct: 648 EPSSWR*VLQYAWTEGCFGISGVSRKFSLHKTRHCSRYCYKSTCISLATNSWTTTRGCFG 827
Query: 720 ESRSFVW*ISLWQCFWGRKWSC*QG*NHK*NSCETWLQLQWERFHLFRYYGLPITSLYFY 779
+ W +C+ + C* +H * + +WL ER + R Y
Sbjct: 828 KGHRLCWFTE--ECYSFLHFLC*---SHY*PAS*SWLFKMGERTSIQRKDR*DGPIANIY 992
Query: 780 GANILPEIKTHGAG*DACSW*WP*SPDN*TANRRES*KWRSTSRRNGT*LLNCIWC 835
G NILP H WP SP + TA R + WR +G *L + WC
Sbjct: 993 GPNILPATAPHVRRQSQIQEHWPSSPADPTACR*QEAIWRYQVW*DGA*LPDSSWC 1160
>AW697009 similar to PIR|D96805|D96 probable acyl-CoA synthetase 62297-59022
[imported] - Arabidopsis thaliana, partial (22%)
Length = 634
Score = 33.9 bits (76), Expect = 0.29
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 6/39 (15%)
Frame = +2
Query: 711 WKDDR--ASWRESRSFVW---*ISLWQCFW-GRKWSC*Q 743
WK R +SWR S +VW *I L CFW G WSC*+
Sbjct: 365 WKKVREASSWRVSMGYVWEGF*ICL*FCFWFGEDWSC*R 481
>CA990983 similar to GP|22202663|dbj putative elicitor response protein
{Oryza sativa (japonica cultivar-group)}, partial (20%)
Length = 444
Score = 29.3 bits (64), Expect = 7.1
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +2
Query: 732 QCFWGRKWSC*QG*N 746
QC WGR+WSC + *N
Sbjct: 239 QCLWGRRWSCLEC*N 283
>TC85925 weakly similar to GP|20259099|gb|AAM14265.1 putative
ubiquitin-conjugating enzyme E2 {Arabidopsis thaliana},
partial (52%)
Length = 3277
Score = 28.9 bits (63), Expect = 9.3
Identities = 7/11 (63%), Positives = 10/11 (90%)
Frame = +3
Query: 731 WQCFWGRKWSC 741
WQC+W ++WSC
Sbjct: 1665 WQCYWFQRWSC 1697
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.365 0.160 0.668
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,822,674
Number of Sequences: 36976
Number of extensions: 632960
Number of successful extensions: 7821
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1678
Number of HSP's successfully gapped in prelim test: 297
Number of HSP's that attempted gapping in prelim test: 5994
Number of HSP's gapped (non-prelim): 2279
length of query: 911
length of database: 9,014,727
effective HSP length: 105
effective length of query: 806
effective length of database: 5,132,247
effective search space: 4136591082
effective search space used: 4136591082
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135796.9


