

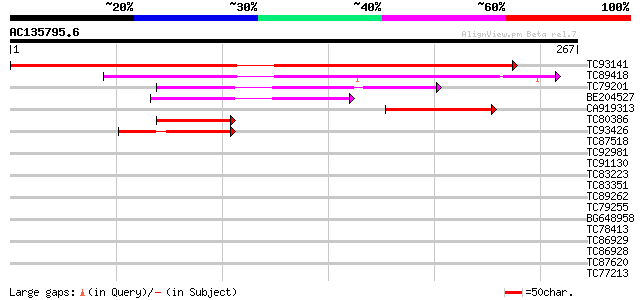
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.6 - phase: 0
(267 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {... 400 e-112
TC89418 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 122 1e-28
TC79201 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 72 2e-13
BE204527 similar to GP|10800070|dbj contains ESTs C61063(AU06336... 57 1e-08
CA919313 56 2e-08
TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 52 3e-07
TC93426 similar to PIR|E84826|E84826 hypothetical protein At2g40... 40 7e-04
TC87518 weakly similar to PIR|E96714|E96714 probable DNA-binding... 40 0.001
TC92981 similar to PIR|T47758|T47758 hypothetical protein F24I3.... 39 0.002
TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidop... 34 0.051
TC83223 similar to GP|8777436|dbj|BAA97026.1 gb|AAC78547.1~gene_... 34 0.067
TC83351 similar to PIR|T47788|T47788 hypothetical protein F17J16... 34 0.067
TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsi... 33 0.11
TC79255 similar to PIR|E96714|E96714 probable DNA-binding protei... 33 0.15
BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidops... 32 0.20
TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 32 0.20
TC86929 similar to PIR|T50498|T50498 myc-like protein - Arabidop... 32 0.33
TC86928 similar to PIR|E96714|E96714 probable DNA-binding protei... 32 0.33
TC87620 similar to GP|22138098|gb|AAM93429.1 ACR4 {Arabidopsis t... 32 0.33
TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding pro... 32 0.33
>TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {Arabidopsis
thaliana}, partial (57%)
Length = 839
Score = 400 bits (1029), Expect = e-112
Identities = 209/239 (87%), Positives = 212/239 (88%)
Frame = +2
Query: 1 MLQQQINNIGMENNSELFQFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVS 60
MLQQQINNIGMENNSELFQFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVS
Sbjct: 173 MLQQQINNIGMENNSELFQFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVS 352
Query: 61 EITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTC 120
EITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKT
Sbjct: 353 EITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKT------------- 493
Query: 121 ERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKAS 180
K S LAKVVERVKEL+QQT QITQ+ETVP E D TVISAGSD+SGEGRLIFKAS
Sbjct: 494 ----DKASLLAKVVERVKELKQQTSQITQLETVPSETDEITVISAGSDISGEGRLIFKAS 661
Query: 181 LCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQN 239
LCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLV AA KEHNSIESIHFLQN
Sbjct: 662 LCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVGAAXKEHNSIESIHFLQN 838
>TC89418 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (26%)
Length = 1093
Score = 122 bits (307), Expect = 1e-28
Identities = 75/230 (32%), Positives = 129/230 (55%), Gaps = 15/230 (6%)
Frame = +3
Query: 45 SSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCN 104
S S + + Y F+ + S +DRA +A K+H +AEKRRR+RIN+ L +LR L+P +
Sbjct: 126 SDSSSGSFQYSEFQSWNLPIEGSVEDRAASASKSHSQAEKRRRDRINTQLANLRKLIPKS 305
Query: 105 SKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTV-- 162
K K + L V+++VK+L+++ +++V TVP E D ++
Sbjct: 306 DKM-----------------DKAALLGSVIDQVKDLKRKAMDVSRVITVPTEIDEVSIDY 434
Query: 163 -------ISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGR 215
+ + +I KAS+CC+DR +L +LI++LKSL L T+KA++A++GGR
Sbjct: 435 NHVVEDETNTNKVNKFKDNIIIKASVCCDDRPELFSELIQVLKSLRLTTVKADIASVGGR 614
Query: 216 TRNVLVVAAEKEHNSIESIHFLQNSLRSLLDR------SSGCNDRSKRRR 259
+++LV+ ++ ++ I+ L+ SL+S + + S C RSKR+R
Sbjct: 615 IKSILVLCSKDSEENV-CINTLKQSLKSAVTKIASSSMVSNCPTRSKRQR 761
>TC79201 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (18%)
Length = 951
Score = 72.4 bits (176), Expect = 2e-13
Identities = 43/134 (32%), Positives = 70/134 (52%)
Frame = +3
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSR 129
++ L ALKNH+EAE++RR RIN HL LR L+P + K K +
Sbjct: 255 NKTLVALKNHREAERKRRNRINGHLAKLRALVPSSPK-----------------MDKATL 383
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDL 189
LA+V+ +VK L++ + ++ ++P + D V G ++KAS+ C+ R +L
Sbjct: 384 LAEVIRQVKHLKKNADEASKGYSIPTDDDEVKV----EPYENGGSFLYKASISCDYRPEL 551
Query: 190 IPDLIEILKSLHLK 203
+ DL + L L L+
Sbjct: 552 LSDLRQTLDKLQLQ 593
>BE204527 similar to GP|10800070|dbj contains ESTs C61063(AU063369)
C61063(AU093310)~unknown protein, partial (18%)
Length = 579
Score = 56.6 bits (135), Expect = 1e-08
Identities = 35/96 (36%), Positives = 50/96 (51%)
Frame = +3
Query: 67 SQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSK 126
S +R++ ALKNH EAE+RRR RIN+HLD LR ++P K K
Sbjct: 342 STTERSVEALKNHSEAERRRRARINAHLDTLRCVIPGALK-----------------MDK 470
Query: 127 GSRLAKVVERVKELRQQTCQITQVETVPCEADGXTV 162
S L +VV +KEL++ Q + +P + D +V
Sbjct: 471 ASLLGEVVRHLKELKRNETQACEGLMIPKDNDEISV 578
>CA919313
Length = 786
Score = 55.8 bits (133), Expect = 2e-08
Identities = 25/52 (48%), Positives = 39/52 (74%)
Frame = -1
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHN 229
+ASLCC+ + DL+ D+ + L +LH +KA++ATLGGR +NV+V+ + KE N
Sbjct: 384 RASLCCQYKPDLLSDIRKALDALHPMIIKAKIATLGGRIKNVVVIISCKEQN 229
>TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (17%)
Length = 655
Score = 51.6 bits (122), Expect = 3e-07
Identities = 23/37 (62%), Positives = 31/37 (83%)
Frame = +1
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSK 106
+R++ ALKNH EAE+RRR RINSHLD LR+++P +K
Sbjct: 499 ERSIEALKNHSEAERRRRARINSHLDTLRSVIPGANK 609
>TC93426 similar to PIR|E84826|E84826 hypothetical protein At2g40200
[imported] - Arabidopsis thaliana, partial (12%)
Length = 637
Score = 40.4 bits (93), Expect = 7e-04
Identities = 22/55 (40%), Positives = 33/55 (60%)
Frame = +1
Query: 52 NYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSK 106
N+ H + ++ QQ A K+H +AEKRRR+RIN+ L +LR L+P + K
Sbjct: 154 NFSHGIYLLKVVLKIEQQ----VASKSHSQAEKRRRDRINTQLANLRKLIPKSDK 306
>TC87518 weakly similar to PIR|E96714|E96714 probable DNA-binding protein
T6L1.19 [imported] - Arabidopsis thaliana, partial (41%)
Length = 1337
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/64 (39%), Positives = 29/64 (45%)
Frame = +2
Query: 45 SSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCN 104
SSS NNNN D P + RA H E E+RRR +IN LR L+P N
Sbjct: 128 SSSINNNNNVK-------VDEPIRGKRANPHRSKHSETEQRRRSKINERFQALRDLIPEN 286
Query: 105 SKTR 108
R
Sbjct: 287 DSKR 298
>TC92981 similar to PIR|T47758|T47758 hypothetical protein F24I3.60 -
Arabidopsis thaliana, partial (27%)
Length = 828
Score = 39.3 bits (90), Expect = 0.002
Identities = 34/126 (26%), Positives = 57/126 (44%), Gaps = 4/126 (3%)
Frame = +3
Query: 19 QFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPF----EVSEITDTPSQQDRALA 74
QF +N E P S + + +S + YH F E EI PS + +A
Sbjct: 42 QFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPSP--KLMA 215
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
NH +E+ RR++INS + LR+LLP +T++ SR + ++ +
Sbjct: 216 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 395
Query: 135 ERVKEL 140
++ +EL
Sbjct: 396 KKKEEL 413
>TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidopsis
thaliana}, partial (20%)
Length = 766
Score = 34.3 bits (77), Expect = 0.051
Identities = 22/69 (31%), Positives = 34/69 (48%)
Frame = +2
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A + H +EKRRR RIN + L+ L+P ++KT K S L + +
Sbjct: 32 AAEVHNLSEKRRRSRINEKMKALQNLIPNSNKT-----------------DKASMLDEAI 160
Query: 135 ERVKELRQQ 143
E +K+L+ Q
Sbjct: 161 EYLKQLQLQ 187
>TC83223 similar to GP|8777436|dbj|BAA97026.1
gb|AAC78547.1~gene_id:MHM17.7~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 514
Score = 33.9 bits (76), Expect = 0.067
Identities = 23/69 (33%), Positives = 36/69 (51%), Gaps = 3/69 (4%)
Frame = +1
Query: 78 NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS---RTCERFKSKGSRLAKVV 134
+H +E+RRRE++N + LR LLP QG K+S E +S + + K+
Sbjct: 199 HHMISERRRREKLNENFQALRALLP------QGTKKDKASILITAKETLRSLMAEIDKLS 360
Query: 135 ERVKELRQQ 143
+R +EL Q
Sbjct: 361 KRNQELMSQ 387
>TC83351 similar to PIR|T47788|T47788 hypothetical protein F17J16.110 -
Arabidopsis thaliana, partial (18%)
Length = 882
Score = 33.9 bits (76), Expect = 0.067
Identities = 19/49 (38%), Positives = 28/49 (56%)
Frame = +1
Query: 59 VSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKT 107
V + T S R A + H +E+RRR+RIN + L+ L+P +SKT
Sbjct: 28 VRKKTSQRSGSARRNRAAEVHNLSERRRRDRINEKMKALQQLIPHSSKT 174
>TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsis
thaliana}, partial (40%)
Length = 1198
Score = 33.1 bits (74), Expect = 0.11
Identities = 27/123 (21%), Positives = 58/123 (46%), Gaps = 1/123 (0%)
Frame = +3
Query: 82 AEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELR 141
AE+RRR+R+N L LR ++P SK + ++ T + K +++E++K L+
Sbjct: 567 AERRRRKRLNDRLSMLRAIVPKISKMDR---TAILGDTIDYMK-------ELLEKIKNLQ 716
Query: 142 QQTCQITQVETVPCEADGXTVISAGS-DMSGEGRLIFKASLCCEDRSDLIPDLIEILKSL 200
Q+ + + ++ + ++ S E + +CC + L+ + L++L
Sbjct: 717 QEIELDSNMTSIVKDVKPNEILIRNSPKFEVERSADTRVEICCAGKPGLLLSTVNTLEAL 896
Query: 201 HLK 203
L+
Sbjct: 897 GLE 905
>TC79255 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (30%)
Length = 847
Score = 32.7 bits (73), Expect = 0.15
Identities = 23/95 (24%), Positives = 37/95 (38%)
Frame = +2
Query: 49 NNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTR 108
N+N V D+A H E+RRR +IN LR L+P
Sbjct: 215 NSNKKQATSSVPNTNKDGKATDKASVIRSKHSVTEQRRRSKINERFQILRDLIP------ 376
Query: 109 QGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQ 143
C++ + S L +V+E V+ L+++
Sbjct: 377 ----------QCDQKRDTASFLLEVIEYVQYLQER 451
>BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidopsis
thaliana}, partial (15%)
Length = 806
Score = 32.3 bits (72), Expect = 0.20
Identities = 29/102 (28%), Positives = 42/102 (40%), Gaps = 15/102 (14%)
Frame = +3
Query: 20 FLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEIT-----DTPSQQDRALA 74
F + NN S Y MQ F ++ N+ SE+ D PS + R
Sbjct: 117 FRLNNNNS--NYQPQRQMPMQIDFSGANSRANSVRPVIAESELVADVEADQPSDERRPRK 290
Query: 75 ALK----------NHKEAEKRRRERINSHLDHLRTLLPCNSK 106
+ NH EAE++RRE++N LR ++P SK
Sbjct: 291 RGRKPANGRDEPLNHVEAERQRREKLNQRFYALRAVVPNISK 416
>TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (30%)
Length = 1397
Score = 32.3 bits (72), Expect = 0.20
Identities = 50/218 (22%), Positives = 89/218 (39%), Gaps = 26/218 (11%)
Frame = +2
Query: 32 STPSSTMMQ-QSFCSSSDNNNNYYHPFEVSEI---------------TDTPSQQD--RAL 73
S+PSST Q SF + + N P +E+ T P Q R++
Sbjct: 338 SSPSSTTSQILSFDCTLNTKKNKVVPLSQTELPQNRKGSLQKQNIVETIKPQGQGTKRSV 517
Query: 74 AALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKV 133
A ++H AE++RRE+++ L L L+P K + +S + K RL +
Sbjct: 518 AHNQDHIIAERKRREKLSQCLIALAALIPGLKKMDK---ASVLGDAIKYVKELQERLRVL 688
Query: 134 VERVKELRQQ---TCQITQVETVPCEADGXTVISAGSDM--SGEGRLIFKASLC---CED 185
E+ K Q T Q+ +D V S ++ E +++ K L C+
Sbjct: 689 EEQNKNSHVQSVVTVDEQQLSYDSSNSDDSEVASGNNETLPHVEAKVLDKDVLIRIHCQK 868
Query: 186 RSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVA 223
+ L+ ++ ++ LHL + + G ++ +VA
Sbjct: 869 QKGLLLKILVEIQKLHLFVVNNSVLPFGDSILDITIVA 982
>TC86929 similar to PIR|T50498|T50498 myc-like protein - Arabidopsis
thaliana, partial (14%)
Length = 1289
Score = 31.6 bits (70), Expect = 0.33
Identities = 26/110 (23%), Positives = 39/110 (34%), Gaps = 8/110 (7%)
Frame = +3
Query: 7 NNIGMENNSELFQFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEI---- 62
N+ G N+ Q N SF E + Q +N ++ E S +
Sbjct: 828 NHHGSFNSRSSSQTTGQKNQSFMEMMKSAQDCAQDE---ELENEGTFFLKRESSNVQRGE 998
Query: 63 ----TDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTR 108
D S + H E+RRR +IN LR L+P + + R
Sbjct: 999 LRVRVDGKSTDQKPNTPRSKHSATEQRRRSKINDRFQMLRELIPHSDQKR 1148
>TC86928 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1926
Score = 31.6 bits (70), Expect = 0.33
Identities = 26/110 (23%), Positives = 39/110 (34%), Gaps = 8/110 (7%)
Frame = +3
Query: 7 NNIGMENNSELFQFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEI---- 62
N+ G N+ Q N SF E + Q +N ++ E S +
Sbjct: 348 NHHGSFNSRSSSQTTGQKNQSFMEMMKSAQDCAQDE---ELENEGTFFLKRESSNVQRGE 518
Query: 63 ----TDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTR 108
D S + H E+RRR +IN LR L+P + + R
Sbjct: 519 LRVRVDGKSTDQKPNTPRSKHSATEQRRRSKINDRFQMLRELIPHSDQKR 668
>TC87620 similar to GP|22138098|gb|AAM93429.1 ACR4 {Arabidopsis thaliana},
partial (91%)
Length = 1508
Score = 31.6 bits (70), Expect = 0.33
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Frame = +2
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEK----EHNSIES 233
K LC DR L+ ++ I + L +AE+ T GG+ N V + +IES
Sbjct: 977 KLELCTTDRVGLLSNVTRIFRENSLTVTRAEVTTKGGKAVNTFYVRGASGCIVDSKTIES 1156
Query: 234 I 234
I
Sbjct: 1157 I 1159
>TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding protein PG1 -
kidney bean, partial (70%)
Length = 2391
Score = 31.6 bits (70), Expect = 0.33
Identities = 17/49 (34%), Positives = 25/49 (50%)
Frame = +3
Query: 78 NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSK 126
NH EAE++RRE++N LR ++P SK + + KSK
Sbjct: 1545 NHVEAERQRREKLNQKFYALRAVVPNVSKMDKASLLGDAISYINELKSK 1691
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,957,358
Number of Sequences: 36976
Number of extensions: 102907
Number of successful extensions: 819
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 810
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 816
length of query: 267
length of database: 9,014,727
effective HSP length: 94
effective length of query: 173
effective length of database: 5,538,983
effective search space: 958244059
effective search space used: 958244059
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135795.6


