

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.1 - phase: 0
(694 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
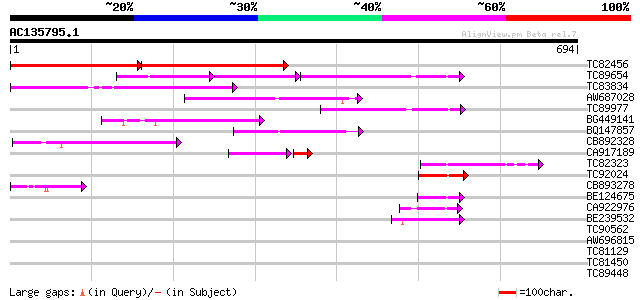
Score E
Sequences producing significant alignments: (bits) Value
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 286 e-129
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 114 4e-65
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 187 9e-48
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 148 6e-36
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 135 4e-32
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 127 2e-29
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 86 6e-17
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 82 5e-16
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 60 5e-11
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 64 2e-10
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 57 2e-08
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 52 1e-06
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 48 1e-05
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 45 9e-05
BE239532 45 1e-04
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 41 0.001
AW696815 homologue to GP|11994736|dbj far-red impaired response ... 36 0.043
TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 35 0.074
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 35 0.074
TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43... 33 0.37
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 286 bits (731), Expect(2) = e-129
Identities = 129/180 (71%), Positives = 149/180 (82%)
Frame = +2
Query: 162 LGTGDAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDT 221
L GD +AI ++F RMQK+NSQFY MD+DDK ++N+FW DARCRAAYEYFGEVIT DT
Sbjct: 704 LWRGDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDT 883
Query: 222 TYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIIT 281
TYLT+KYD+P PFVGVNHHGQ++LLGCA+LSN D KT +WLF WLECMHG APN IIT
Sbjct: 884 TYLTSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIIT 1063
Query: 282 DQDRAMKKAIEDVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDF 341
++D+AMK AIE FPKARHRWCLWH+MKKVPE LG++S ESIK LL D VYDS S SDF
Sbjct: 1064EEDKAMKNAIEVAFPKARHRWCLWHIMKKVPEMLGKYSRNESIKTLLQDVVYDSMSKSDF 1243
Score = 194 bits (492), Expect(2) = e-129
Identities = 94/162 (58%), Positives = 116/162 (71%)
Frame = +1
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKPN 60
MTF SEEEVI+YY +YA +GFG KI+SKNA DGKKYFTL C AR YVS SKN K
Sbjct: 220 MTFRSEEEVIRYYMNYANRVGFGVTKISSKNADDGKKYFTLACNCARRYVSTSKNPSKQY 399
Query: 61 PTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIND 120
T + QC AR+N ++LDG TI+++ LEHNH L TK RYFR +++ H +R L++ND
Sbjct: 400 LTSKTQCRARLNACVALDGSSTISRIVLEHNHELGQTKGRYFRLDESSGTHVERNLELND 579
Query: 121 QAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRL 162
Q GINV RN +S+ +E NG NLT GE DCRN++ K RRLRL
Sbjct: 580 QGGINVDRNLQSVGLEMNGCGNLTSGENDCRNFVQKVRRLRL 705
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 114 bits (286), Expect(3) = 4e-65
Identities = 64/200 (32%), Positives = 103/200 (51%)
Frame = +1
Query: 357 NEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNA 416
NEW + +++ R WVPVY+R +F+ + +E +N FFDG+V++ TTL+ V QY+ A
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 417 LKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEG 476
+ E+E ADF + N+ + E Q +T F +FQ E+ + A
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFMKFQEELVGTMANPATKIE 1056
Query: 477 MENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLT 536
+T+ V + + T V FN + + C+C +FE+ GI+CRHIL V +
Sbjct: 1057DSGTITTYRVAKFGE-----NQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFR-A 1218
Query: 537 GKTESVPSCYILSRWRKDIK 556
++PS Y+L RW + K
Sbjct: 1219KNVLTLPSHYVLKRWTMNAK 1278
Score = 99.4 bits (246), Expect(3) = 4e-65
Identities = 48/122 (39%), Positives = 69/122 (56%), Gaps = 2/122 (1%)
Frame = +2
Query: 131 RSMVVEANGYDNLTFGEKDCR--NYIDKARRLRLGTGDAEAIQNYFVRMQKKNSQFYYVM 188
R + V+ +L+ C+ ++ R+ LG G + + +Y RM+ +N F+Y +
Sbjct: 11 RFLCVDYRVISSLSINPNCCKL*AHMSSTRQRTLGGGGHQVL-DYLKRMRAENHAFFYAV 187
Query: 189 DVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLG 248
D + N+FW D CR Y YFG+ + FDTTY TN+Y +PFA F G NHHGQ VL G
Sbjct: 188 QSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTNQYRVPFASFTGFNHHGQPVLFG 367
Query: 249 CA 250
CA
Sbjct: 368 CA 373
Score = 74.3 bits (181), Expect(3) = 4e-65
Identities = 36/111 (32%), Positives = 58/111 (51%), Gaps = 1/111 (0%)
Frame = +3
Query: 247 LGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKARHRWCLWH 306
L L+ NE ++ WLFKTWL + GR P +I TD D ++ A+ V P RHR+C W
Sbjct: 363 LAVLLILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAVAQVLPPTRHRFCKWS 542
Query: 307 LMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHD 356
+ ++ KL + + V++S +I +F W+ ++E Y + D
Sbjct: 543 IFRENRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLERYYIMD 695
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 187 bits (476), Expect = 9e-48
Identities = 103/280 (36%), Positives = 156/280 (54%), Gaps = 1/280 (0%)
Frame = +3
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGK-KYFTLGCTRARSYVSNSKNLLKP 59
M F S ++ YY YA+ +GF NS ++ K KY + C ++ + +K++
Sbjct: 225 MEFESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEKYGAVLCCSSQGF-KRTKDVNNL 401
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIN 119
R C A + M + + I +V LEHNH L + +K++ K L +
Sbjct: 402 RKETRTGCPAMIRMKLVESQRWRICEVTLEHNHVLGA------KIHKSIK---KNSLPSS 554
Query: 120 DQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQK 179
D G + + + ++V++ G DNL +D R + + +L L GD +AI N+ RMQ
Sbjct: 555 DAEGKTI-KVYHALVIDTEGNDNLNSNARDDRAFSKYSNKLNLRKGDTQAIYNFLCRMQL 731
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVN 239
N F+Y+MD +D+ LRN WVDA+ RAA YF +VI FD TYL NKY++P VG+N
Sbjct: 732 TNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYLVNKYEIPLVALVGIN 911
Query: 240 HHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAI 279
HHGQSVLLGC LL+ E +++ WLF+TW++C+ +P I
Sbjct: 912 HHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 148 bits (374), Expect = 6e-36
Identities = 77/225 (34%), Positives = 119/225 (52%), Gaps = 8/225 (3%)
Frame = +1
Query: 215 EVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGR 274
+V+ FDTTY NKY+ P F G NHH Q+++ G ALL +E +++ W+ +LECM +
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 275 APNAIITDQDRAMKKAIEDVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYD 334
P A++TD D +M++AI+ VFP A HR C WHL K E + + E + A+Y
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFR----KAMYS 348
Query: 335 SSSISDFMEKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNS 394
+ + F + W ++I+ EL N W+ + + W Y+RD F+ + TT + E++N+
Sbjct: 349 NFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAINA 528
Query: 395 FFDGYVSSKTT--------LKQFVEQYDNALKDKIEKESMADFVS 431
Y S+ L +F E Y N E +ADF S
Sbjct: 529 IVKTYSRSQRQNF*IYA*FLNRF*EGYRN-------NELVADFKS 642
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 135 bits (341), Expect = 4e-32
Identities = 78/178 (43%), Positives = 103/178 (57%)
Frame = +2
Query: 381 AGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACIS 440
AG ST QRSESMNSFFD Y+ K TLK+FV+QY L ++ ++E +ADF + + A S
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 441 LFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTR 500
+E Q +T+A FK+FQ+E+ + C + +E + + V D KD
Sbjct: 182 PSPWEKQMSTIYTHAIFKKFQVEVLGVAGCQSRIEVGDGTAVRYI------VQDYEKDEE 343
Query: 501 FRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRR 558
F V + E E+ C C LFE+KG LCRH L VLQ G SVPS YI+ RW KD K R
Sbjct: 344 FLVTWKELSSEVSCFCRLFEYKGFLCRHALSVLQRCG-CSSVPSHYIMKRWTKDAKIR 514
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 127 bits (318), Expect = 2e-29
Identities = 71/207 (34%), Positives = 111/207 (53%), Gaps = 7/207 (3%)
Frame = +3
Query: 113 KRRLDINDQAGINVSRNFRSMVVEA---NGYDNLTFGEKDCRNYIDKARRLRLGTGDAEA 169
K R+ + + GI+V + R M +E GY L F EKD RN + R+L D E
Sbjct: 72 KNRILMFAKTGISVHQMMRLMELEKCVEPGY--LPFTEKDVRNLLQSFRKL-----DPEE 230
Query: 170 IQNYFVRM----QKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLT 225
+RM + K+ F + +D +RL N+ W A Y+ FG+ + FDTT+
Sbjct: 231 ETLDLLRMCRNIKDKDPNFKFEYTLDANNRLENIAWSYASSIQLYDIFGDAVVFDTTHRL 410
Query: 226 NKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDR 285
+DMP +VG+N++G GC LL +E ++FSW K +L M+G+AP I+TDQ+
Sbjct: 411 TAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSFSWAIKAFLGFMNGKAPQTILTDQNI 590
Query: 286 AMKKAIEDVFPKARHRWCLWHLMKKVP 312
+K+A+ P +H +C+W ++ K P
Sbjct: 591 CLKEALSAEMPMTKHAFCIWMIVAKFP 671
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 85.5 bits (210), Expect = 6e-17
Identities = 46/161 (28%), Positives = 88/161 (54%), Gaps = 2/161 (1%)
Frame = +1
Query: 275 APNAIITDQDRAMKKAIEDVFPKARHRWCLWHLMKKVPE--KLGRHSHYESIKLLLHDAV 332
AP ++TD + +K+AI P+++H +C+WH++ K + L S Y+ K H +
Sbjct: 1 APQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFH-RL 177
Query: 333 YDSSSISDFMEKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESM 392
Y+ DF + W++M++ Y LH N+ + L+ R W ++R F+AG+++ ++ES+
Sbjct: 178 YNLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESI 357
Query: 393 NSFFDGYVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFN 433
N F ++S+++ +F++Q +AD V FN
Sbjct: 358 NVFIQRFLSAQSQPXRFLQQ-------------VADIVDFN 441
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 82.4 bits (202), Expect = 5e-16
Identities = 57/217 (26%), Positives = 95/217 (43%), Gaps = 10/217 (4%)
Frame = +1
Query: 4 SSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKPNPT- 62
+S++E Y+ +A GF K + KK TR + + + + P
Sbjct: 175 NSKDEAYDLYQEHAFKTGFSVRKGKELYYDNEKKK-----TRLKDFYCSKQGFKNNEPDG 339
Query: 63 --------IRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYF-RCNKNLDPHTK 113
R C+A V +++ +G +TK+ L+HNH P + RY R +N+ +
Sbjct: 340 EVAYKRADSRTNCLAMVRFNVTKEGVWKVTKLILDHNHEFVPLQQRYLLRSMRNMSNLKE 519
Query: 114 RRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNY 173
R+ I V+ + E G D KD NY+ + + GDA+++ N+
Sbjct: 520 DRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMHNYVSTGKLKFIEAGDAQSLLNH 699
Query: 174 FVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAY 210
Q ++S FYY + +D +SRL NVFW D + Y
Sbjct: 700 LQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKSVVDY 810
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 59.7 bits (143), Expect(2) = 5e-11
Identities = 30/79 (37%), Positives = 41/79 (50%), Gaps = 1/79 (1%)
Frame = +1
Query: 268 LECMHGRAPNAIITDQDRAMKKAIEDVFPKARHRWCLWHLMKKVPEKLGR-HSHYESIKL 326
L M GR P +I TD D ++ AI VFP RHR+C WH+ K+ EKL + + +
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 327 LLHDAVYDSSSISDFMEKW 345
H V + SI +F W
Sbjct: 181 EFHKCVNLTDSIDEFESCW 237
Score = 26.2 bits (56), Expect(2) = 5e-11
Identities = 8/23 (34%), Positives = 15/23 (64%)
Frame = +3
Query: 348 MIECYELHDNEWLKGLFDERYRW 370
+++ Y+L DNEWL+ + +W
Sbjct: 243 LLDRYDLRDNEWLQAIHSACRQW 311
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 63.9 bits (154), Expect = 2e-10
Identities = 44/153 (28%), Positives = 70/153 (44%), Gaps = 3/153 (1%)
Frame = +1
Query: 504 IFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRR--HTL 561
++N E QC+C LFE +GI C H+ V++ + +P C +++RW K+ K ++
Sbjct: 1 VYNAASLEFQCSCRLFESRGIPCCHVFFVMK-EEDVDHIPKCLVMTRWTKNAKGEFLNSD 177
Query: 562 IKCGFDGN-VELQLVGKACDAFYEVVSVGIHTEDDLLKVMNRINELKIELNCEGSSSVII 620
D N +EL G C AF +M+ I L+ + C+ S
Sbjct: 178 SNGEIDANMIELARFGAYCSAFTTFCKEASKKNGVFRDIMDEILNLQ-KKYCDVEDST-- 348
Query: 621 EEDSLVQNQMAKILDPATTRSKGRPPSKRKASK 653
S +Q+ DP T +SKG P K+ A+K
Sbjct: 349 RSKSFTDDQVG---DPVTVKSKGAPKKKKNAAK 438
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 57.4 bits (137), Expect = 2e-08
Identities = 27/61 (44%), Positives = 39/61 (63%)
Frame = +3
Query: 501 FRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRRHT 560
+ V FN + + C+C +FEF G+LCRHIL V ++T ++PS YIL RW K+ K +
Sbjct: 54 YNVRFNVLEMKATCSCQMFEFSGLLCRHILAVFRVT-NVLTLPSHYILKRWTKNAKSNVS 230
Query: 561 L 561
L
Sbjct: 231 L 233
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 51.6 bits (122), Expect = 1e-06
Identities = 34/102 (33%), Positives = 50/102 (48%), Gaps = 8/102 (7%)
Frame = +3
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLG----CT----RARSYVSN 52
M F+S EE +Y Y R +GF TV+I+ N + +G C+ RA+ YV
Sbjct: 327 MQFNSREEARGFYDGYGRRIGF-TVRIHH-NRRSRVNNELIGQDFVCSKEGFRAKKYVHR 500
Query: 53 SKNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGL 94
+L P P R C A + +++ +GK +TK EH H L
Sbjct: 501 KDRVLPPPPATREGCQAMIRLALRDEGKWVVTKFVKEHTHKL 626
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 47.8 bits (112), Expect = 1e-05
Identities = 24/57 (42%), Positives = 31/57 (54%)
Frame = +2
Query: 500 RFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIK 556
++ V F+ D + C+C FE G LC H L VL + VPS YIL RW KD +
Sbjct: 245 QYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLD-NRNIKVVPSRYILKRWTKDAR 412
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 45.1 bits (105), Expect = 9e-05
Identities = 27/77 (35%), Positives = 41/77 (53%)
Frame = -3
Query: 478 ENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
E N ++ + SKK+ D V++ + +I C+C FE GILCRH L +L +T
Sbjct: 795 ELYNGSYIIRHSKKL-----DGERHVLWLPEKEQILCSCKEFESSGILCRHALRIL-VTK 634
Query: 538 KTESVPSCYILSRWRKD 554
+P Y LSRW ++
Sbjct: 633 NYFQLPEKYYLSRWHRE 583
>BE239532
Length = 645
Score = 44.7 bits (104), Expect = 1e-04
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 5/94 (5%)
Frame = +3
Query: 468 IYCNAFLEGMEN-----LNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFK 522
I+ N +LEG ++ + + + + + + ++ V F+ + +I C+C F+
Sbjct: 87 IFLNEYLEGTGGSTSIEISQSNNLSNHEVMLNHKPNKKYAVTFDSSNMKINCSCRKFDSM 266
Query: 523 GILCRHILCVLQLTGKTESVPSCYILSRWRKDIK 556
GILC H L + + G +P Y L RW K+ +
Sbjct: 267 GILCSHALRIYNIKGILR-IPDQYFLKRWSKNAR 365
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/102 (29%), Positives = 50/102 (48%), Gaps = 4/102 (3%)
Frame = +2
Query: 3 FSSEEEVIKYYKSYARCMGFGTVKIN--SKNAKDGKKYF-TLGCTRARSYVSNSKN-LLK 58
F SE E +Y SYA +GF ++++ S++ +DG L C + + + + +++
Sbjct: 341 FVSEAEAHAFYNSYATRVGF-VIRVSKLSRSRRDGSVIGRALVCNKEGFRMPDKREKIVR 517
Query: 59 PNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSR 100
R C A + + G +ITK EH H L+P KSR
Sbjct: 518 QRAETRVGCRAMIMVRKLNSGLWSITKFVKEHTHPLTPGKSR 643
>AW696815 homologue to GP|11994736|dbj far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (7%)
Length = 394
Score = 36.2 bits (82), Expect = 0.043
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Frame = +2
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYF--TLGCTR 45
M F S E +Y+ YAR MGF T NS+ +K +++ C+R
Sbjct: 227 MEFESHGEAYSFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSR 367
>TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (13%)
Length = 721
Score = 35.4 bits (80), Expect = 0.074
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Frame = +2
Query: 3 FSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYF--TLGCTRARSYVSNSKNLLKPN 60
F + E +Y+ YA+ MGF T NS+ +K K++ C+R + + +
Sbjct: 431 FETHEAAYSFYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGVTPESDGRSSQRS 610
Query: 61 PTIRAQCMARVNMSMSLDGKITI 83
+ C A +++ + DGK I
Sbjct: 611 SVKKTDCKACMHVKRNPDGKWNI 679
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 35.4 bits (80), Expect = 0.074
Identities = 34/130 (26%), Positives = 52/130 (39%), Gaps = 4/130 (3%)
Frame = +2
Query: 4 SSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVSNSKN--LLKPNP 61
SS +++ + S R +V S++ KDG Y K +++
Sbjct: 62 SSSACILQ*HXSSQRLDSSYSVSKLSRSRKDGTAIGRALVCNKEGYRMPDKREKIVRQRA 241
Query: 62 TIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSR--YFRCNKNLDPHTKRRLDIN 119
R C A + M GK ITK EH H L+P SR F + + H K R +++
Sbjct: 242 ETRVGCRAMIMMRKINSGKWVITKFVKEHTHPLNPGSSRRDMFEIYPSQNEHDKIR-ELS 418
Query: 120 DQAGINVSRN 129
Q I R+
Sbjct: 419 QQLAIEKKRS 448
>TC89448 similar to PIR|C84864|C84864 hypothetical protein At2g43280
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1177
Score = 33.1 bits (74), Expect = 0.37
Identities = 27/90 (30%), Positives = 42/90 (46%), Gaps = 6/90 (6%)
Frame = +1
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINS--KNAKDGKKYF-TLGCTRARSYVSNSKNL- 56
M F SE+ +Y YAR +GF +++ S ++ +DG+ GC + VS L
Sbjct: 313 MEFVSEDAAKIFYDEYARHVGF-VMRVMSCRRSXRDGRILARRFGCNKEGHCVSIQGKLG 489
Query: 57 --LKPNPTIRAQCMARVNMSMSLDGKITIT 84
KP + R C A +++ GK IT
Sbjct: 490 PVRKPRASTREGCKAMIHIKYDQSGKWMIT 579
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,346,733
Number of Sequences: 36976
Number of extensions: 325492
Number of successful extensions: 1817
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 1795
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1805
length of query: 694
length of database: 9,014,727
effective HSP length: 103
effective length of query: 591
effective length of database: 5,206,199
effective search space: 3076863609
effective search space used: 3076863609
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135795.1


