

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135565.15 + phase: 0
(557 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
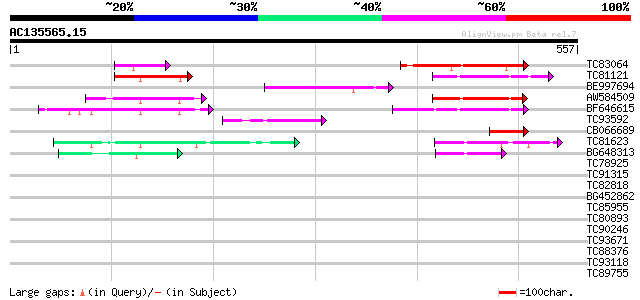
Score E
Sequences producing significant alignments: (bits) Value
TC83064 similar to GP|14423452|gb|AAK62408.1 Unknown protein {Ar... 150 1e-36
TC81121 homologue to GP|18182311|gb|AAL65125.1 GT-2 factor {Glyc... 84 1e-16
BE997694 similar to GP|13646986|db DNA-binding protein DF1 {Pisu... 80 2e-15
AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pi... 80 2e-15
BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabido... 75 9e-14
TC93592 weakly similar to GP|13646986|dbj|BAB41080. DNA-binding ... 64 1e-10
CB066689 homologue to GP|13646986|dbj DNA-binding protein DF1 {P... 62 4e-10
TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcri... 59 5e-09
BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Ara... 52 5e-07
TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Ar... 39 0.007
TC91315 weakly similar to GP|8777369|dbj|BAA96959.1 gene_id:MJE7... 39 0.007
TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Ar... 37 0.026
BG452862 weakly similar to GP|14715258|emb N3 like protein {Medi... 36 0.044
TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {... 36 0.044
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 36 0.044
TC90246 similar to PIR|T00872|T00872 probable protein kinase At2... 36 0.044
TC93671 weakly similar to PIR|F84595|F84595 hypothetical protein... 36 0.044
TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidop... 35 0.099
TC93118 homologue to GP|14587596|gb|AAK70588.1 NADH dehydrogenas... 35 0.099
TC89755 similar to GP|10177798|dbj|BAB11289. emb|CAB87778.1~gene... 34 0.13
>TC83064 similar to GP|14423452|gb|AAK62408.1 Unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 571
Score = 150 bits (379), Expect = 1e-36
Identities = 81/133 (60%), Positives = 96/133 (71%), Gaps = 8/133 (6%)
Frame = +1
Query: 385 PSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDEVLALINLRC-----NNNNEEK 439
PSSS T +N S+ + + E+ + RRWP+DEVLALINL+ N +N
Sbjct: 37 PSSSDT-----EYSNSTSTLVVPTIMEKLEDRRRWPRDEVLALINLKSTTSVINRSNNNV 201
Query: 440 EGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDA---NRKRSLDSRT 496
EGNSN K PLWERIS+GM ELGYKRSAKRCKEKWENINKYF+KTKD +KRS+DSRT
Sbjct: 202 EGNSN-KGPLWERISEGMFELGYKRSAKRCKEKWENINKYFKKTKDIVVNKKKRSMDSRT 378
Query: 497 CPYFHLLTNLYNQ 509
CPYFH L++LYNQ
Sbjct: 379 CPYFHQLSSLYNQ 417
Score = 48.1 bits (113), Expect = 9e-06
Identities = 24/66 (36%), Positives = 38/66 (57%), Gaps = 11/66 (16%)
Frame = +1
Query: 104 WTNDEVLALLKIRSSME-----------SWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFE 152
W DEVLAL+ ++S+ + WE +S + E+GYKRSA++CKEK+E
Sbjct: 124 WPRDEVLALINLKSTTSVINRSNNNVEGNSNKGPLWERISEGMFELGYKRSAKRCKEKWE 303
Query: 153 EESRFF 158
+++F
Sbjct: 304 NINKYF 321
>TC81121 homologue to GP|18182311|gb|AAL65125.1 GT-2 factor {Glycine max},
partial (37%)
Length = 794
Score = 84.0 bits (206), Expect = 1e-16
Identities = 44/119 (36%), Positives = 70/119 (57%)
Frame = +3
Query: 416 GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWEN 475
G RWP+ E LAL+ +R + + ++ S+ K PLWE +S+ + +LGY RS+K+CKEK+EN
Sbjct: 288 GNRWPRQETLALLKIRSDMDGVFRD--SSLKGPLWEEVSRKLADLGYHRSSKKCKEKFEN 461
Query: 476 INKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQ 534
+ KY ++TK+ +S + +T +F L L Q L S + N N P N ++
Sbjct: 462 VYKYHKRTKEGRSGKS-EGKTYRFFDQLQALEKQ--LTFSSYHPKPQTNNNTPTTNPIE 629
Score = 71.6 bits (174), Expect = 7e-13
Identities = 38/83 (45%), Positives = 51/83 (60%), Gaps = 7/83 (8%)
Frame = +3
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W E LALLKIRS M+ F D + WE VSRKLA++GY RS++KCKEKFE ++
Sbjct: 297 WPRQETLALLKIRSDMDGVFRDSSLKGPLWEEVSRKLADLGYHRSSKKCKEKFENVYKYH 476
Query: 159 NNINHNQN--SFGKNFRFVTELE 179
++ S GK +RF +L+
Sbjct: 477 KRTKEGRSGKSEGKTYRFFDQLQ 545
>BE997694 similar to GP|13646986|db DNA-binding protein DF1 {Pisum sativum},
partial (19%)
Length = 609
Score = 80.5 bits (197), Expect = 2e-15
Identities = 42/135 (31%), Positives = 78/135 (57%), Gaps = 8/135 (5%)
Frame = +3
Query: 251 FKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQ 310
+K F E ++K+++++QEE+H + +E + KR+ E+ +REEAW+ QEM+++N+E E++A E+
Sbjct: 9 WKDFFERLMKEVVEKQEELHKRFLEAIEKRERERGAREEAWRLQEMQRINREREILAQER 188
Query: 311 AIAGDRQAHIIQFLNKFSTSANSSSL--------TSMSTQLQAYLATLTSNSSSSTLHSQ 362
++A + A ++ FL K + +L T + Q QA T+ + + T +
Sbjct: 189 SLAATKDAAVMAFLQKIAEQQEQQNLVPPVLNNSTIVPQQQQAPQETIPTPTPKPT-PTP 365
Query: 363 NPNPETLKKTLQPIP 377
P P L P+P
Sbjct: 366 TPTPVPLPAAAAPLP 410
>AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pisum
sativum}, partial (22%)
Length = 694
Score = 80.1 bits (196), Expect = 2e-15
Identities = 38/93 (40%), Positives = 61/93 (64%)
Frame = +2
Query: 416 GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWEN 475
G RWP+ E LAL+ +R + + K+ + K PLW+ +S+ M +LGY+R++K+CKEK+EN
Sbjct: 188 GNRWPRQETLALLKIRSDMDGAFKDASV--KGPLWDEVSRKMADLGYQRNSKKCKEKFEN 361
Query: 476 INKYFRKTKDANRKRSLDSRTCPYFHLLTNLYN 508
+ KY ++TK+ +S D +T +F L L N
Sbjct: 362 VYKYHKRTKEGRGGKS-DGKTYRFFDQLQALEN 457
Score = 70.5 bits (171), Expect = 2e-12
Identities = 44/126 (34%), Positives = 66/126 (51%), Gaps = 7/126 (5%)
Frame = +2
Query: 75 DKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFPDFT-----WE 129
D E+ S N +DR + W E LALLKIRS M+ F D + W+
Sbjct: 128 DDERGGGSSRNEEGVDRSFGG------NRWPRQETLALLKIRSDMDGAFKDASVKGPLWD 289
Query: 130 HVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQ--NSFGKNFRFVTELEEVYQGGGG 187
VSRK+A++GY+R+++KCKEKFE ++ + S GK +RF +L+ +
Sbjct: 290 EVSRKMADLGYQRNSKKCKEKFENVYKYHKRTKEGRGGKSDGKTYRFFDQLQAL------ 451
Query: 188 ENNKNL 193
ENN ++
Sbjct: 452 ENNPSM 469
>BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabidopsis
thaliana, partial (16%)
Length = 654
Score = 74.7 bits (182), Expect = 9e-14
Identities = 43/134 (32%), Positives = 74/134 (55%), Gaps = 1/134 (0%)
Frame = +1
Query: 377 PENPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDEVLALINLRCNNNN 436
P ++ P+++TT + P + + + + GR WP+ E L L+ +R +
Sbjct: 226 PAVATTATPTNTTTSASTPSFSGNLEAETTAFIAVDASTGR-WPRQETLTLLEIRSRLDP 402
Query: 437 EEKEGNSNNKAPLWERISQGMLEL-GYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSR 495
+ KE + K PLW+++S+ M E GY+RS K+C+EK+EN+ KY++KTK+ R D +
Sbjct: 403 QFKE--ATQKGPLWDQLSRIMCEEHGYQRSGKKCREKFENLYKYYKKTKEGKAGRQ-DGK 573
Query: 496 TCPYFHLLTNLYNQ 509
+F L LY +
Sbjct: 574 HYRFFRQLEALYGE 615
Score = 65.5 bits (158), Expect = 5e-11
Identities = 57/196 (29%), Positives = 87/196 (44%), Gaps = 24/196 (12%)
Frame = +1
Query: 29 PLSTPNNTFPPFDPYNQQNHPSQHHQLPL------QVQPNLLHPL----HPHKDDEDKEQ 78
P ST NN FP + + N QH+++ + + P LH P
Sbjct: 79 PRSTTNN-FPTAELFPAGNPHQQHYEMMMFGRQVADIIPRCLHDFVSTDSPAVATTATPT 255
Query: 79 N-----STPSMN-NFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFPDFT----- 127
N STPS + N + + + W E L LL+IRS ++ F + T
Sbjct: 256 NTTTSASTPSFSGNLEAETTAFIAVDASTGRWPRQETLTLLEIRSRLDPQFKEATQKGPL 435
Query: 128 WEHVSRKLAEV-GYKRSAEKCKEKFEEESRFFNNINHNQ--NSFGKNFRFVTELEEVYQG 184
W+ +SR + E GY+RS +KC+EKFE +++ + GK++RF +LE +Y
Sbjct: 436 WDQLSRIMCEEHGYQRSGKKCREKFENLYKYYKKTKEGKAGRQDGKHYRFFRQLEALY-- 609
Query: 185 GGGENNKNLVEAEKQN 200
GENN + QN
Sbjct: 610 --GENNNPSSSSLPQN 651
>TC93592 weakly similar to GP|13646986|dbj|BAB41080. DNA-binding protein DF1
{Pisum sativum}, partial (11%)
Length = 622
Score = 64.3 bits (155), Expect = 1e-10
Identities = 35/102 (34%), Positives = 60/102 (58%)
Frame = +1
Query: 210 EEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEM 269
E S DDV S S +E +KR+R + E F ++VKKM ++QE+M
Sbjct: 250 EPGSSSDDVGNSSASTKEF-------GSRKRRRKSSRKLE---DFAANLVKKMTEKQEQM 399
Query: 270 HNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQA 311
H +++E + + ++E+ REEAWK++EME++ ++ E A E++
Sbjct: 400 HKEMVEMIERMEKERIKREEAWKREEMERIKQDEEARAAERS 525
>CB066689 homologue to GP|13646986|dbj DNA-binding protein DF1 {Pisum
sativum}, partial (8%)
Length = 508
Score = 62.4 bits (150), Expect = 4e-10
Identities = 25/38 (65%), Positives = 31/38 (80%)
Frame = +3
Query: 472 KWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQ 509
KWENINKYF+K K++N+KR DS+TCPYFH L LY +
Sbjct: 3 KWENINKYFKKVKESNKKRPEDSKTCPYFHQLDALYKE 116
>TC81623 similar to PIR|T01257|T01257 probable GT-1-like transcription
factor [imported] - Arabidopsis thaliana, partial (47%)
Length = 847
Score = 58.9 bits (141), Expect = 5e-09
Identities = 59/259 (22%), Positives = 105/259 (39%), Gaps = 18/259 (6%)
Frame = +1
Query: 44 NQQNHPSQHHQLPLQVQPNLLHPLHPHKDDEDKEQN--STPSMNNFQIDRDQRQILPQLI 101
+ + H QHHQ Q H H + + N T S+N DR PQ
Sbjct: 136 SMEGHHLQHHQQHQHQQHQQHHQQHQQRQHQQHAHNIIGTSSVNVDVTDR-----FPQ-- 294
Query: 102 DPWTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEE-ES 155
W+ E L IRS ++ F + WE +S + E GY RSAE+CK K++ +
Sbjct: 295 --WSIQETKEFLMIRSELDQTFMETKRNKQLWEVISNTMKEKGYHRSAEQCKCKWKNLVT 468
Query: 156 RFFNNINHNQNSFGKNFRFVTELEEVY----------QGGGGENNKNLVEAEKQNEVQDK 205
R+ + + F F EL+ ++ + GG K + +E +++
Sbjct: 469 RYKGCETMEVEAMRQQFPFYNELQAIFSARMQRMLWAEAEGGSKKKGV---HVSSEDEEE 639
Query: 206 MDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQ 265
+ E + + KK + +++ G ND +G + + ++++ M Q
Sbjct: 640 LGNEESEGDHKGNIKKKKKKGKMIIGGGGND-----NNGSNNL-------KEILEEFMRQ 783
Query: 266 QEEMHNKLIEDMVKRDEEK 284
Q +M + +E R+ E+
Sbjct: 784 QMQMEAQWMEAFEARENER 840
Score = 42.7 bits (99), Expect = 4e-04
Identities = 33/131 (25%), Positives = 64/131 (48%), Gaps = 5/131 (3%)
Frame = +1
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENIN 477
+W E + +R + E N + LWE IS M E GY RSA++CK KW+N+
Sbjct: 292 QWSIQETKEFLMIRSELDQTFMETKRNKQ--LWEVISNTMKEKGYHRSAEQCKCKWKNLV 465
Query: 478 KYFR--KTKDANRKRSLDSRTCPYFHLLTNLYN---QGKLVLQSDQKQESNNVNVPEENV 532
++ +T + R + P+++ L +++ Q L +++ + V+V E+
Sbjct: 466 TRYKGCETMEVEAMR----QQFPFYNELQAIFSARMQRMLWAEAEGGSKKKGVHVSSED- 630
Query: 533 VQEKAKQDENQ 543
+E+ +E++
Sbjct: 631 -EEELGNEESE 660
>BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Arabidopsis
thaliana}, partial (53%)
Length = 815
Score = 52.4 bits (124), Expect = 5e-07
Identities = 28/70 (40%), Positives = 38/70 (54%)
Frame = +1
Query: 419 WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINK 478
W +DE +LI LR ++ SN LWE+IS M E G+ RS C +KW N+ K
Sbjct: 160 WVQDETRSLIGLRREMDSLFNTSKSNKH--LWEQISAKMREKGFDRSPTMCTDKWRNLLK 333
Query: 479 YFRKTKDANR 488
F+K K +R
Sbjct: 334 EFKKAKHHDR 363
Score = 49.3 bits (116), Expect = 4e-06
Identities = 32/126 (25%), Positives = 49/126 (38%), Gaps = 5/126 (3%)
Frame = +1
Query: 49 PSQHHQLPLQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDE 108
PSQHH Q QP ++ DD + E + + + W DE
Sbjct: 43 PSQHHHHLQQQQPQMILTAESSGDDPEMEIKAP----------------KKRAETWVQDE 174
Query: 109 VLALLKIRSSMESWF-----PDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINH 163
+L+ +R M+S F WE +S K+ E G+ RS C +K+ + F H
Sbjct: 175 TRSLIGLRREMDSLFNTSKSNKHLWEQISAKMREKGFDRSPTMCTDKWRNLLKEFKKAKH 354
Query: 164 NQNSFG 169
+ G
Sbjct: 355 HDRGGG 372
>TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 1274
Score = 38.5 bits (88), Expect = 0.007
Identities = 18/72 (25%), Positives = 35/72 (48%), Gaps = 4/72 (5%)
Frame = +1
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNN----KAPLWERISQGMLELGYKRSAKRCKEKW 473
RW + E+L LI + + + K G + + P W +S + G R +C+++W
Sbjct: 211 RWTRQEILVLIQGKSDAESRFKPGRNGSGFGSSEPKWALVSSYCKKHGVNRGPIQCRKRW 390
Query: 474 ENINKYFRKTKD 485
N+ ++K K+
Sbjct: 391 SNLAGDYKKIKE 426
>TC91315 weakly similar to GP|8777369|dbj|BAA96959.1
gene_id:MJE7.2~pir||C71412~similar to unknown protein
{Arabidopsis thaliana}, partial (3%)
Length = 1044
Score = 38.5 bits (88), Expect = 0.007
Identities = 17/58 (29%), Positives = 33/58 (56%)
Frame = +2
Query: 434 NNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRS 491
N + ++ N NNK + E+++Q LEL ++ A K +W+ I ++F + A K++
Sbjct: 113 NMGDVEQSNENNKESMIEQLAQAFLELEAQKGASEDKVQWDEIKQHFSDLEMALNKKN 286
>TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1176
Score = 36.6 bits (83), Expect = 0.026
Identities = 13/36 (36%), Positives = 26/36 (72%)
Frame = +1
Query: 450 WERISQGMLELGYKRSAKRCKEKWENINKYFRKTKD 485
W+ IS+ M E GY+ S ++C++K+ ++NK +++ D
Sbjct: 7 WKSISKVMAERGYRVSPQQCEDKFNDLNKRYKRLND 114
Score = 34.3 bits (77), Expect = 0.13
Identities = 11/35 (31%), Positives = 25/35 (71%)
Frame = +1
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNIN 162
W+ +S+ +AE GY+ S ++C++KF + ++ + +N
Sbjct: 7 WKSISKVMAERGYRVSPQQCEDKFNDLNKRYKRLN 111
>BG452862 weakly similar to GP|14715258|emb N3 like protein {Medicago
truncatula}, partial (33%)
Length = 658
Score = 35.8 bits (81), Expect = 0.044
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 2/89 (2%)
Frame = +3
Query: 202 VQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKK 261
+ +K+ H+ D +V K+ E VV TTNDEKK + S D +V+K
Sbjct: 363 IDEKLPEHKGD------IVDKEIENVVVPSKTTNDEKKLEVSVVDMV---------IVEK 497
Query: 262 MMDQQEEMHN--KLIEDMVKRDEEKFSRE 288
++Q+E H+ + +D V +D+ K E
Sbjct: 498 KEEKQDEEHDEKEKKQDQVTQDKTKVKNE 584
>TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {Cicer
arietinum}, partial (98%)
Length = 1981
Score = 35.8 bits (81), Expect = 0.044
Identities = 36/158 (22%), Positives = 63/158 (39%)
Frame = +2
Query: 136 AEVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVE 195
+E G EK +E+ EE F++ + KN + E E V
Sbjct: 1250 SEKGTSAFGEKLREQVEERLDFYDKGVAPR----KNIDVMKSAIESVDNKDTEMETEEVS 1417
Query: 196 AEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFC 255
A+K + + K + DD+ V K +E + G D K K+
Sbjct: 1418 AKKTKKKKQKA------ADGDDMAVDKAAE---ITNGDAEDHKSEKKK------------ 1534
Query: 256 ESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKK 293
+ K+ +DQ+ E+ +K++ED D K ++++ KK
Sbjct: 1535 KKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 1648
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 35.8 bits (81), Expect = 0.044
Identities = 33/129 (25%), Positives = 60/129 (45%), Gaps = 17/129 (13%)
Frame = -1
Query: 318 AHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNP-----ETLKKT 372
+H + F + S++SSS +S S+ + ++ + +SSSS+ S P P + +
Sbjct: 595 SHFLFFAGGWGVSSSSSSSSSSSSSSPSSSSSSSPSSSSSSPSSLPPAPLASGSSSGSPS 416
Query: 373 LQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLI------------SSGERDDIGRRWP 420
P SS+ PSSS++ + +++ SS SL+ S G + I +
Sbjct: 415 SSPSSPE*SSSSPSSSSSCTSSSSSSSSSSSVSLLFSL*AGFLPRRCSKGNLESIAVNFG 236
Query: 421 KDEVLALIN 429
E+ ALI+
Sbjct: 235 ATELAALIS 209
>TC90246 similar to PIR|T00872|T00872 probable protein kinase At2g45590
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1451
Score = 35.8 bits (81), Expect = 0.044
Identities = 25/92 (27%), Positives = 43/92 (46%)
Frame = +2
Query: 211 EDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMH 270
+D MD + KSE+ +E G +EKK+++ + E ++ E +V MM
Sbjct: 194 KDYVMDWIGREVKSEKSELEGGKIGNEKKKEKKRKQKLEWWESMDEEIVGGMM------- 352
Query: 271 NKLIEDMVKRDEEKFSREEAWKKQEMEKMNKE 302
R E++ S E WK++ E++ KE
Sbjct: 353 ---------RKEKRKSVREWWKEEHCEEVAKE 421
Score = 28.9 bits (63), Expect = 5.4
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 1/71 (1%)
Frame = -1
Query: 319 HIIQFLNKFSTSANS-SSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIP 377
H FL+ FS S +S S L + + + + T +S SS+ H Q P P ++ IP
Sbjct: 470 HHHHFLHLFSFSYSSFSPLQPLHNAPPSTIPSPTFSSLSSSSHQQFPRP-----SIPTIP 306
Query: 378 ENPSSTLPSSS 388
+ SS+ P SS
Sbjct: 305 ISASSSSPFSS 273
>TC93671 weakly similar to PIR|F84595|F84595 hypothetical protein At2g20980
[imported] - Arabidopsis thaliana, partial (42%)
Length = 938
Score = 35.8 bits (81), Expect = 0.044
Identities = 21/41 (51%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Frame = +3
Query: 364 PNPETLKKTLQPIPENPSSTLPSSSTTLV-AQPRNNNPISS 403
PNP TL TL P+P NPS T+P+ + +LV A P N P+ S
Sbjct: 189 PNPLTLTLTLNPLP-NPSPTIPNLTNSLVFALPTNV*PLLS 308
>TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidopsis
thaliana}, partial (25%)
Length = 706
Score = 34.7 bits (78), Expect = 0.099
Identities = 19/66 (28%), Positives = 33/66 (49%)
Frame = +1
Query: 335 SLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQ 394
+L++MS + + L T T + + NP ++ TL P+P P++ P S + Q
Sbjct: 94 ALSNMSLEARNLLQTTTQPNLPNIPTLPNPTLPSIPTTLPPLPSIPTTLPPLPSMPTLPQ 273
Query: 395 PRNNNP 400
P+ N P
Sbjct: 274 PQGNVP 291
>TC93118 homologue to GP|14587596|gb|AAK70588.1 NADH dehydrogenase subunit 3
{Microcebus rufus}, partial (10%)
Length = 567
Score = 34.7 bits (78), Expect = 0.099
Identities = 23/68 (33%), Positives = 34/68 (49%), Gaps = 1/68 (1%)
Frame = +3
Query: 332 NSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTL 391
N S S +Q +Y+A+LT N + + H+QNP + ENP+ TL + + L
Sbjct: 135 NGDSPLSPRSQWFSYIASLTQNQNQNNNHNQNPFMSQI--------ENPNQTLEQAFSRL 290
Query: 392 -VAQPRNN 398
VA P N
Sbjct: 291 SVANPNPN 314
>TC89755 similar to GP|10177798|dbj|BAB11289.
emb|CAB87778.1~gene_id:MXA21.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (59%)
Length = 864
Score = 34.3 bits (77), Expect = 0.13
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 7/52 (13%)
Frame = +1
Query: 327 FSTSANSSSLTSMSTQLQAYLAT-------LTSNSSSSTLHSQNPNPETLKK 371
FS+S++SSS S S LQ ++T +++N+S +T HS NPN + +
Sbjct: 16 FSSSSSSSSSYSYSYSLQNQISTKKKTHRIISTNTSRATFHSPNPNQKNFTR 171
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.127 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,474,819
Number of Sequences: 36976
Number of extensions: 283532
Number of successful extensions: 2758
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 2544
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2680
length of query: 557
length of database: 9,014,727
effective HSP length: 101
effective length of query: 456
effective length of database: 5,280,151
effective search space: 2407748856
effective search space used: 2407748856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135565.15


