

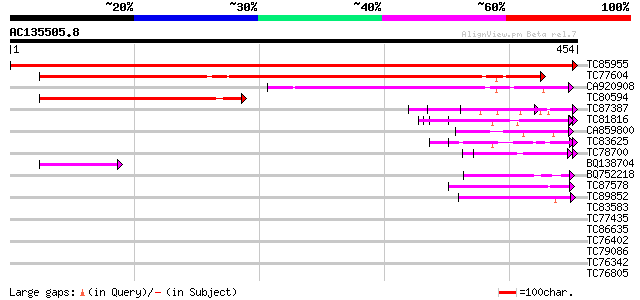
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {... 889 0.0
TC77604 homologue to PIR|T06377|T06377 SAR DNA-binding protein-1... 303 9e-83
CA920908 homologue to PIR|T06377|T06 SAR DNA-binding protein-1 -... 140 8e-34
TC80594 homologue to PIR|T06377|T06377 SAR DNA-binding protein-1... 135 2e-32
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 60 2e-09
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 57 1e-08
CA859800 weakly similar to GP|17473819|gb| putative protein {Ara... 51 8e-07
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 47 1e-05
TC78700 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsi... 46 3e-05
BQ138704 similar to PIR|T06377|T06 SAR DNA-binding protein-1 - g... 45 8e-05
BQ752218 similar to GP|19170914|emb hypothetical protein {Enceph... 42 5e-04
TC87578 similar to PIR|T12113|T12113 transcription factor - fava... 42 6e-04
TC89852 similar to GP|15620063|gb|AAL03490.1 unknown {Rickettsia... 41 8e-04
TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 -... 40 0.001
TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna... 40 0.002
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 39 0.003
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 38 0.007
TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein l... 38 0.009
TC76342 similar to PIR|A86444|A86444 probable RNA helicase [impo... 37 0.012
TC76805 similar to PIR|T17104|T17104 translation initiation fact... 37 0.021
>TC85955 similar to GP|3860319|emb|CAA10127.1 nucleolar protein {Cicer
arietinum}, partial (98%)
Length = 1981
Score = 889 bits (2296), Expect = 0.0
Identities = 454/454 (100%), Positives = 454/454 (100%)
Frame = +2
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ
Sbjct: 296 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 475
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM
Sbjct: 476 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 655
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA
Sbjct: 656 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 835
Query: 181 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 240
AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR
Sbjct: 836 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 1015
Query: 241 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 300
LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR
Sbjct: 1016LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 1195
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 360
MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES
Sbjct: 1196MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 1375
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK
Sbjct: 1376VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 1555
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE
Sbjct: 1556LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 1657
>TC77604 homologue to PIR|T06377|T06377 SAR DNA-binding protein-1 - garden
pea, partial (85%)
Length = 1625
Score = 303 bits (776), Expect = 9e-83
Identities = 177/411 (43%), Positives = 257/411 (62%), Gaps = 6/411 (1%)
Frame = +3
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+LGVA+SK+G+ I E KI N V EL+RGVR ++ + L D+ LGL HS
Sbjct: 390 TLGVADSKLGNIIKEKLKIECVHNNAVMELMRGVRYQLNELIAGLAVQDMAPMSLGLSHS 569
Query: 85 YSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYC 144
SR ++KF+ ++VD M++QAI LLD LDK++N++AMRVREWY WHFPEL KI+ DN Y
Sbjct: 570 LSRYRLKFSADKVDTMIVQAIGLLDDLDKELNTYAMRVREWYGWHFPELTKIIVDNIQYA 749
Query: 145 KVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRV 204
+ K + ++ A+ ++++ +E +A E+ EA+ SMG ++ +DL N+ +V
Sbjct: 750 RSVKLMGNRINAAK---LDFSEILSEEVEA-EVKEASVISMGTEIGELDLSNIRELCDQV 917
Query: 205 MDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGA 264
+ LS+YR +L DYL ++MN IAPNL ++VG+ VGARLI+H GSL NLAK P ST+QILGA
Sbjct: 918 LSLSEYRAQLYDYLKSRMNTIAPNLTAMVGELVGARLIAHGGSLINLAKQPGSTVQILGA 1097
Query: 265 EKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTS 324
EKALFRALKT+ TPKYGLI+H+S IG+A+ + KG+++R LA K ++A R D + +
Sbjct: 1098EKALFRALKTKHATPKYGLIYHASLIGQAAPKFKGKISRSLAAKTALAIRCDALGDGQDN 1277
Query: 325 AFGEKLREQVEERL-DFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQ 383
G + R ++E RL + K + K IE+ D + ++ KT
Sbjct: 1278TMGLENRAKLEARLRNLEGKELGRFAGSAKGKPKIEAYDKDRKKGAGGLITPAKT---YN 1448
Query: 384 KAADG-----DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVED 429
AAD + A+D+ + D K EKK+KK++K+K +E EVED
Sbjct: 1449TAADSVIEPMSNSAMDE--DTPEPPVTDKKKEKKEKKEKKKKEKKEEEVED 1595
>CA920908 homologue to PIR|T06377|T06 SAR DNA-binding protein-1 - garden pea,
partial (37%)
Length = 748
Score = 140 bits (354), Expect = 8e-34
Identities = 99/255 (38%), Positives = 145/255 (56%), Gaps = 10/255 (3%)
Frame = -2
Query: 207 LSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAEK 266
LS+YR +L D + P G+ VGARLI+H GSL NLAK P ST+QILGAEK
Sbjct: 747 LSEYRAQLYDI*RVG*IPLHP-FDCYGGELVGARLIAHGGSLINLAKQPGSTVQILGAEK 571
Query: 267 ALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSAF 326
ALFRALKT+ TPKYGLI+H+S IG+A+ + KG+++R LA K ++A R D + +
Sbjct: 570 ALFRALKTKHATPKYGLIYHASLIGQAAPKFKGKISRSLAAKTALAIRCDALGDGQDNTM 391
Query: 327 GEKLREQVEERL-DFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKA 385
G + R ++E RL + K + K IE+ D + ++ KT A
Sbjct: 390 GLENRAKLEARLRNLEGKELGRFAGSAKGKPKIEAYDKDRKKGAGGLITPAKTYNT---A 220
Query: 386 ADG-----DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVE----VEDKVVEDGA 436
AD D A+D+ E + A D K EKK+KKK+++K +++ + D+ ++ +
Sbjct: 219 ADSIIDKKSDSAMDEDTEELS--AADKKKEKKEKKKKEKKEEEDTQKSNTAMDEDTQEPS 46
Query: 437 NADSSKKKKKKSKKK 451
AD K+KK+K +KK
Sbjct: 45 AADKKKEKKEKKEKK 1
>TC80594 homologue to PIR|T06377|T06377 SAR DNA-binding protein-1 - garden
pea, partial (45%)
Length = 907
Score = 135 bits (341), Expect = 2e-32
Identities = 73/166 (43%), Positives = 104/166 (61%), Gaps = 1/166 (0%)
Frame = +2
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+L VA++K+G+ I E KI + V E++RG+R + + L D+ LGL HS
Sbjct: 347 TLAVADTKLGTIIKEKLKIDCVHSNAVMEIMRGIRYQLTELITGLAVQDMAPMSLGLSHS 526
Query: 85 YSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYC 144
SR K+KF+ ++VD M++QAI LLD LDK++N++AMRVREWY WHFPEL KI+ DN Y
Sbjct: 527 LSRYKLKFSADKVDTMIVQAIGLLDDLDKELNTYAMRVREWYGWHFPELTKIIQDNIQYA 706
Query: 145 KVAKFIEDKSKLAE-DKIESLTDLVGDEDKAKEIVEAAKASMGQDL 189
+ K + D+ A+ D E LT E+ E+ EAA SMG ++
Sbjct: 707 RSVKLMGDRINAAKLDFSEILT-----EEVEAELKEAAVISMGTEI 829
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 60.1 bits (144), Expect = 2e-09
Identities = 39/100 (39%), Positives = 52/100 (52%), Gaps = 7/100 (7%)
Frame = +3
Query: 362 DNKDTEMETEEVSAKKTKKKKQKAADGD------DMAVDKAAEITN-GDAEDHKSEKKKK 414
+N + E+V +K KK K+K DG D K E + G E KSEKKKK
Sbjct: 420 ENGAAASDEEKVEKEKKKKHKEKGEDGSPEVEKSDKKKKKHKETSEVGSPEVDKSEKKKK 599
Query: 415 KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KK+K D ++ + D +NAD S+KK KK K KDA+
Sbjct: 600 KKDKEAKDNAADISNG--NDESNADRSEKKHKKKKNKDAQ 713
Score = 44.3 bits (103), Expect = 1e-04
Identities = 34/96 (35%), Positives = 48/96 (49%), Gaps = 6/96 (6%)
Frame = +3
Query: 335 EERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSA----KKTKKKKQKAADGDD 390
EE+++ K K D +S K ET EV + K KKKK+K D
Sbjct: 444 EEKVEKEKKKKHKEKGEDGSPEVEKSDKKKKKHKETSEVGSPEVDKSEKKKKKK----DK 611
Query: 391 MAVDKAAEITNGDAEDH--KSEKKKKKKEKRKLDQE 424
A D AA+I+NG+ E + +SEKK KKK+ + +E
Sbjct: 612 EAKDNAADISNGNDESNADRSEKKHKKKKNKDAQEE 719
Score = 43.9 bits (102), Expect = 1e-04
Identities = 45/146 (30%), Positives = 62/146 (41%), Gaps = 11/146 (7%)
Frame = +3
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMK-SAIESVDNKDTEMETEEVSAKKT 378
EK GE + + E D AP K V + E V K + EV K
Sbjct: 222 EKVKKDDGEGRKRKKHESAD----SPAPAKKSKVAEVDGEEKVKTKKVDDAAVEVEDDKK 389
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ---EVEVEDK----- 430
+KKK+K D ++ A +++ K EK+KKKK K K + EVE DK
Sbjct: 390 EKKKKKKKDKENGAAA---------SDEEKVEKEKKKKHKEKGEDGSPEVEKSDKKKKKH 542
Query: 431 --VVEDGANADSSKKKKKKSKKKDAE 454
E G+ +KKKK K K+A+
Sbjct: 543 KETSEVGSPEVDKSEKKKKKKDKEAK 620
Score = 37.4 bits (85), Expect = 0.012
Identities = 33/115 (28%), Positives = 47/115 (40%), Gaps = 6/115 (5%)
Frame = +3
Query: 345 VAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKK------KKQKAADGDDMAVDKAAE 398
V P V+ + +D + E + +K KK K++K D A K ++
Sbjct: 129 VLPTGGDAVIAGMAAATGTEDVKKEVADEEGEKVKKDDGEGRKRKKHESADSPAPAKKSK 308
Query: 399 ITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
+ D E+ K K K+ D VEVED E KKKKKK K+ A
Sbjct: 309 VAEVDGEE-------KVKTKKVDDAAVEVEDDKKE-------KKKKKKKDKENGA 431
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 57.4 bits (137), Expect = 1e-08
Identities = 36/124 (29%), Positives = 65/124 (52%)
Frame = +3
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD 387
+K +E+ EE + ++ +K+ + K + +D + + EE +KK K+KK+ +
Sbjct: 258 KKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNE 437
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
DD D + + N K +K+KKK+E K + +V V D +E+ A KKKKK+
Sbjct: 438 DDDEGEDGSKKKKN------KDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGKEKKKKKE 599
Query: 448 SKKK 451
K++
Sbjct: 600 DKEE 611
Score = 47.4 bits (111), Expect = 1e-05
Identities = 39/126 (30%), Positives = 59/126 (45%), Gaps = 10/126 (7%)
Frame = +3
Query: 337 RLDFYDKGVAPRKNIDVMKSA-IESVDNKDTEMETEEVSAKKTKKKKQKA--------AD 387
++D G +++ K I+SV+ D + E KK K+KK K D
Sbjct: 90 KIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKE------KKDKEKKDKTDVDEGKDKKD 251
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSK-KKKK 446
+ +K E G+ ED +K K+KK+K K ++ E +DK DG S K K+KK
Sbjct: 252 KEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDK---DGEEKKSKKDKEKK 422
Query: 447 KSKKKD 452
K K +D
Sbjct: 423 KDKNED 440
Score = 46.6 bits (109), Expect = 2e-05
Identities = 35/123 (28%), Positives = 56/123 (45%), Gaps = 3/123 (2%)
Frame = +3
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDM 391
E+ +E+ + DK + ++D E D KD E + +E + K +++ + D
Sbjct: 168 EKDDEKKEKKDKEKKDKTDVD------EGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDK 329
Query: 392 AVDKAAEITNGDAE---DHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKS 448
K + G + D + +K KK KEK+K DK +D D SKKKK K
Sbjct: 330 EKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKK--------DKNEDDDEGEDGSKKKKNKD 485
Query: 449 KKK 451
KK+
Sbjct: 486 KKE 494
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/103 (32%), Positives = 53/103 (51%)
Frame = +3
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK 411
DV + +E KD E+++ E +K +KK ++ D D+ D K +K
Sbjct: 114 DVKEDKVEI--EKDLEIKSVEKDDEKKEKKDKEKKDKTDV-----------DEGKDKKDK 254
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+KKKKEK+ ++ V+ E+ EDG +KKKK+ K+K E
Sbjct: 255 EKKKKEKK--EENVKGEE---EDGDEKKDKEKKKKEKKEKGKE 368
>CA859800 weakly similar to GP|17473819|gb| putative protein {Arabidopsis
thaliana}, partial (8%)
Length = 531
Score = 51.2 bits (121), Expect = 8e-07
Identities = 37/108 (34%), Positives = 48/108 (44%), Gaps = 14/108 (12%)
Frame = +2
Query: 358 IESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSE------- 410
+E ++ D T E S KK KKKK+K A TNG + +E
Sbjct: 77 VEEIEKNDPTSLTPEASTKKKKKKKKKT---------NTAATTNGTTNSNVTEYTETIEN 229
Query: 411 -KKKKKKEKRKLDQEVEVEDKVVED------GANADSSKKKKKKSKKK 451
KK + E + D EVE++ E+ G A KKKKKKSKKK
Sbjct: 230 IKKTETTETAETDNSKEVEEENEEEEGTEAVGGEASKKKKKKKKSKKK 373
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 47.4 bits (111), Expect = 1e-05
Identities = 39/126 (30%), Positives = 59/126 (45%), Gaps = 10/126 (7%)
Frame = +2
Query: 337 RLDFYDKGVAPRKNIDVMKSA-IESVDNKDTEMETEEVSAKKTKKKKQKA--------AD 387
++D G +++ K I+SV+ D + E KK K+KK K D
Sbjct: 482 KIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKE------KKDKEKKDKTDVDEGKDKKD 643
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSK-KKKK 446
+ +K E G+ ED +K K+KK+K K ++ E +DK DG S K K+KK
Sbjct: 644 KEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDK---DGEEKKSKKDKEKK 814
Query: 447 KSKKKD 452
K K +D
Sbjct: 815 KDKNED 832
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/103 (32%), Positives = 53/103 (51%)
Frame = +2
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK 411
DV + +E KD E+++ E +K +KK ++ D D+ D K +K
Sbjct: 506 DVKEDKVEI--EKDLEIKSVEKDDEKKEKKDKEKKDKTDV-----------DEGKDKKDK 646
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+KKKKEK+ ++ V+ E+ EDG +KKKK+ K+K E
Sbjct: 647 EKKKKEKK--EENVKGEE---EDGDEKKDKEKKKKEKKEKGKE 760
>TC78700 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsis
thaliana}, partial (5%)
Length = 687
Score = 46.2 bits (108), Expect = 3e-05
Identities = 26/79 (32%), Positives = 44/79 (54%)
Frame = +2
Query: 372 EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKV 431
EVS KK KKKK+K+ G++ +K + D KKKKK+++ ++++E E +
Sbjct: 392 EVSEKKEKKKKKKSGSGEEEEKEKEVNLDGEGVGD----KKKKKRKRDEVEKEWEEKKYG 559
Query: 432 VEDGANADSSKKKKKKSKK 450
+ +G + D S +K KK
Sbjct: 560 IVEGGDKDESVEKVGNXKK 616
Score = 42.0 bits (97), Expect = 5e-04
Identities = 29/92 (31%), Positives = 43/92 (46%)
Frame = +2
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLD 422
N+ E V KK KK K+K+ D + + +E +K+KKKK+K
Sbjct: 296 NQTLSPENGVVDEKKRKKNKEKSPVIDSNPIIEVSE-----------KKEKKKKKKSGSG 442
Query: 423 QEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+E E E +V DG KKKK+K + + E
Sbjct: 443 EEEEKEKEVNLDGEGVGDKKKKKRKRDEVEKE 538
>BQ138704 similar to PIR|T06377|T06 SAR DNA-binding protein-1 - garden pea,
partial (26%)
Length = 667
Score = 44.7 bits (104), Expect = 8e-05
Identities = 24/66 (36%), Positives = 37/66 (55%)
Frame = +1
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+L VA++K+G+ I E KI + V E++RG+R + + L D+ LGL HS
Sbjct: 469 TLAVADTKLGTIIKEKLKIDCVHSNAVMEIMRGIRYQLTELITGLAVQDMAXMSLGLSHS 648
Query: 85 YSRAKV 90
SR K+
Sbjct: 649 LSRYKL 666
>BQ752218 similar to GP|19170914|emb hypothetical protein {Encephalitozoon
cuniculi}, partial (5%)
Length = 637
Score = 42.0 bits (97), Expect = 5e-04
Identities = 28/89 (31%), Positives = 45/89 (50%)
Frame = -2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + EE + K+ +KKK+KA + KA E A++ K E +KK KE+++
Sbjct: 633 KKRPKKKEEKAKKEAEKKKKKAREEAAKKAKKAREAAYKKAKEEKKEAEKKAKEEKR--- 463
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ E K+ E+ KK+ +K KKD
Sbjct: 462 --QAEKKIKEE-------KKEAEKKAKKD 403
>TC87578 similar to PIR|T12113|T12113 transcription factor - fava bean,
partial (49%)
Length = 1021
Score = 41.6 bits (96), Expect = 6e-04
Identities = 26/101 (25%), Positives = 45/101 (43%)
Frame = +2
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK 411
D + +E + N+ E E++E +K + + DD D + +GD ++ ++K
Sbjct: 479 DAVDPHLERIRNEAGEDESDEEDEDFVAEKDDEGSPTDDSGADDSDASQSGDEKEIPAKK 658
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ KK K + +AD KKKK+ KKKD
Sbjct: 659 EPKKDLSSKASASTSTSTSK-KKSKDADEDGKKKKQKKKKD 778
Score = 31.6 bits (70), Expect = 0.66
Identities = 22/93 (23%), Positives = 42/93 (44%), Gaps = 3/93 (3%)
Frame = +2
Query: 328 EKLREQV-EERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEM--ETEEVSAKKTKKKKQK 384
E++R + E+ D D+ K+ + + D+ D + +E+ AKK KK
Sbjct: 500 ERIRNEAGEDESDEEDEDFVAEKDDEGSPTDDSGADDSDASQSGDEKEIPAKKEPKKDLS 679
Query: 385 AADGDDMAVDKAAEITNGDAEDHKSEKKKKKKE 417
+ + + + + ED K +K+KKKK+
Sbjct: 680 SKASASTSTSTSKKKSKDADEDGKKKKQKKKKD 778
>TC89852 similar to GP|15620063|gb|AAL03490.1 unknown {Rickettsia conorii},
partial (22%)
Length = 1001
Score = 41.2 bits (95), Expect = 8e-04
Identities = 33/101 (32%), Positives = 50/101 (48%), Gaps = 7/101 (6%)
Frame = +1
Query: 360 SVDNK-DTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDA-EDHKSEKKKKKKE 417
S+D K + ETE+ S K KKK + ++GD + TN +A + K + KK+K+
Sbjct: 646 SIDAKVENGAETEKRSKHKMKKKDKSISEGDAKEQIGDPDATNEEAIPEEKKDSKKRKRP 825
Query: 418 KRKLDQEVEVEDKVVEDG-----ANADSSKKKKKKSKKKDA 453
K + E + E KV E+ N SK ++ SK K A
Sbjct: 826 ISKENDEQDTEMKVDEETKRRKIENGSESKGRRPISKTKRA 948
Score = 37.4 bits (85), Expect = 0.012
Identities = 31/108 (28%), Positives = 50/108 (45%), Gaps = 17/108 (15%)
Frame = +1
Query: 360 SVDNK-DTEMETEEVSAKKTKKKKQKAADG--------------DDMAVDKAAEITNGDA 404
S D K D +ETE+ S K KK + +G DD+++D A++ NG
Sbjct: 505 STDAKVDNGVETEKRSKHKKKKNNKSNIEGGAIEQNAVTENKVKDDLSID--AKVENGAE 678
Query: 405 EDHKSEKKKKKKEKR--KLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
+ +S+ K KKK+K + D + ++ D + KK KK K+
Sbjct: 679 TEKRSKHKMKKKDKSISEGDAKEQIGDPDATNEEAIPEEKKDSKKRKR 822
>TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 - garden
pea, partial (33%)
Length = 852
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/48 (50%), Positives = 33/48 (68%)
Frame = +2
Query: 406 DHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
D K EKK+KK++K+K +E EVED VE+ + KK+KK KKKD+
Sbjct: 398 DKKKEKKEKKEKKKKEKKEEEVED--VEE--PEEEVVKKEKKKKKKDS 529
Score = 38.1 bits (87), Expect = 0.007
Identities = 24/77 (31%), Positives = 39/77 (50%)
Frame = +3
Query: 376 KKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDG 435
+K +++K+K + + AE+ NGD + EKKKK+K++ + ED
Sbjct: 498 RKKRRRKRKI-------LXEKAEVQNGDHTSNGGEKKKKRKKQAE-----------EEDS 623
Query: 436 ANADSSKKKKKKSKKKD 452
A SKKK KK K+ +
Sbjct: 624 AEMPPSKKKDKKKKRAE 674
Score = 37.0 bits (84), Expect = 0.016
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 11/92 (11%)
Frame = +2
Query: 337 RLDFYDKG--------VAPRKNIDVMK-SAIESVDNKDTEMETEE--VSAKKTKKKKQKA 385
+++ YDK + P K + S IE + N + +T E V+ KK +KK++K
Sbjct: 251 KIEAYDKDRKKGAGGLITPAKTYNTAADSVIEPMSNSAMDEDTPEPPVTDKKKEKKEKKE 430
Query: 386 ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKE 417
+ ++ ++ + E K EKKKKKK+
Sbjct: 431 KKKKEKKEEEVEDVEEPEEEVVKKEKKKKKKD 526
>TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna radiata},
partial (67%)
Length = 1828
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/86 (29%), Positives = 43/86 (49%)
Frame = +1
Query: 369 ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
+T + + K+K K+K +D+ ++ +A K ++ KK++K D E
Sbjct: 139 DTVSMESPKSKSSKKKKLLQNDV-------VSEVEAVSAKKKESSKKRKKSSDDDEETKS 297
Query: 429 DKVVEDGANADSSKKKKKKSKKKDAE 454
D E+G+ +KKKKKSK +D E
Sbjct: 298 DISSEEGSRKVKKEKKKKKSKVEDEE 375
Score = 38.5 bits (88), Expect = 0.005
Identities = 27/79 (34%), Positives = 40/79 (50%), Gaps = 2/79 (2%)
Frame = +1
Query: 365 DTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQE 424
D E E VSAKK + K++ DD K+ + + K EKKKKK + D+E
Sbjct: 202 DVVSEVEAVSAKKKESSKKRKKSSDDDEETKSDISSEEGSRKVKKEKKKKKSKVE--DEE 375
Query: 425 VEVEDKVV--EDGANADSS 441
+ E++V +D NA S+
Sbjct: 376 IVEEEEVAVKKDDPNAVSN 432
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 39.3 bits (90), Expect = 0.003
Identities = 26/77 (33%), Positives = 44/77 (56%), Gaps = 5/77 (6%)
Frame = +2
Query: 359 ESVDNKDTEMETEEV----SAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK 414
E DN+ +E ETEE +KT+ + ++ + D+ +DK+ E AE K EK K
Sbjct: 53 EGDDNEKSEEETEEKEEGDEKEKTEAETEEEEEADEDTIDKSKE--EDKAEGSKGEKGSK 226
Query: 415 KKEKRKLDQE-VEVEDK 430
K+ + K+++E V+V+ K
Sbjct: 227 KRARGKVNEEKVKVKKK 277
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 38.1 bits (87), Expect = 0.007
Identities = 33/133 (24%), Positives = 56/133 (41%), Gaps = 16/133 (12%)
Frame = +3
Query: 335 EERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGD-DMAV 393
E++ ++ AP K + +E+ + E +E K K KK+ +D D D
Sbjct: 696 EKKKKIMERKEAPLK----WEQKLEAAAKAKADAEAKEKKLKTAKHKKRSGSDTDSDRDS 863
Query: 394 D---------------KAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
D K + I +GD E K + K K +KR + D E +++
Sbjct: 864 DDERKRASKRSHRKHRKHSHIDSGDHEKRKEKSSKWKTKKRSSESSDFSSD---ESESSS 1034
Query: 439 DSSKKKKKKSKKK 451
+ K++KKK +KK
Sbjct: 1035EEEKRRKKKQRKK 1073
Score = 33.5 bits (75), Expect = 0.17
Identities = 21/91 (23%), Positives = 45/91 (49%)
Frame = +3
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
+D+ D E E+ S KTKK+ +++D + ++E + +++KKK+ K+
Sbjct: 924 IDSGDHEKRKEKSSKWKTKKRSSESSDFSSDESESSSE---------EEKRRKKKQRKKI 1076
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
DQ+ + AD K+K++ +++
Sbjct: 1077 RDQDSRSNSS--GSDSYADEVNKRKRRHRRR 1163
>TC79086 similar to GP|23499033|emb|CAD51113. ubiquitin-protein ligase 1
putative {Plasmodium falciparum 3D7}, partial (0%)
Length = 1418
Score = 37.7 bits (86), Expect = 0.009
Identities = 32/136 (23%), Positives = 64/136 (46%)
Frame = +2
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
SE G+S+ G E+ EE + P + +++E D ++T+ + +E KT
Sbjct: 143 SEAGSSSEGG---EEEEESSEEEAPKTTPPPKTNPKNNSVEDEDEEETDTDEDE-PPPKT 310
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
K +Q G ++ A T AE++ +++ KKK + +++ E E E+ N+
Sbjct: 311 KPIEQTLKSGTKPSIAPARSGTKRPAENNDTKQSNKKKTTEEKNKKKEKEP---EEDDNS 481
Query: 439 DSSKKKKKKSKKKDAE 454
++ K ++ +D E
Sbjct: 482 NNKKASFQRVFTEDDE 529
>TC76342 similar to PIR|A86444|A86444 probable RNA helicase [imported] -
Arabidopsis thaliana, partial (80%)
Length = 1651
Score = 37.4 bits (85), Expect = 0.012
Identities = 35/104 (33%), Positives = 50/104 (47%), Gaps = 9/104 (8%)
Frame = +3
Query: 348 RKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDH 407
RKN V+ + E N +TE+++ S KKTKK K + + K + TN D
Sbjct: 66 RKNDAVLSTEPEQALNNNTELKS---SKKKTKKNKHE----ETTPKRKHQDETNLQ-NDT 221
Query: 408 KSEKKKKKKEKRKLDQE------VEV---EDKVVEDGANADSSK 442
+SEKK KKK+K +E V V ++ +V G NA K
Sbjct: 222 ESEKKSKKKKKHNKSEENNDAGGVSVSASDEPIVVTGKNAGDEK 353
Score = 29.3 bits (64), Expect = 3.3
Identities = 15/48 (31%), Positives = 25/48 (51%)
Frame = +3
Query: 404 AEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
A ++ +E K KK+ +K E + +D N + + +KKSKKK
Sbjct: 105 ALNNNTELKSSKKKTKKNKHEETTPKRKHQDETNLQNDTESEKKSKKK 248
>TC76805 similar to PIR|T17104|T17104 translation initiation factor eIF-2
beta chain - apple tree (fragment), partial (81%)
Length = 1064
Score = 36.6 bits (83), Expect = 0.021
Identities = 27/72 (37%), Positives = 36/72 (49%), Gaps = 8/72 (11%)
Frame = +1
Query: 376 KKTKKKKQKAADGDDMAVDKAAE------ITNGDAEDHKSEKKKKKK--EKRKLDQEVEV 427
KK KKKK D DD +VDK AE ++ G KKKKKK E L E +
Sbjct: 121 KKKKKKKPAVVDLDDDSVDKLAEKTENLSVSEGFDSILAGSKKKKKKPVEISSLIDESDA 300
Query: 428 EDKVVEDGANAD 439
+++ ++D A D
Sbjct: 301 KNEDLDDHAEED 336
Score = 30.0 bits (66), Expect = 1.9
Identities = 23/72 (31%), Positives = 31/72 (42%), Gaps = 6/72 (8%)
Frame = +1
Query: 389 DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA------DSSK 442
D+ +D E+ D KKKKKK+ +D + + DK+ E N DS
Sbjct: 61 DEAQIDVKEEVP--DIAPFDPTKKKKKKKPAVVDLDDDSVDKLAEKTENLSVSEGFDSIL 234
Query: 443 KKKKKSKKKDAE 454
KK KKK E
Sbjct: 235 AGSKKKKKKPVE 270
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,229,882
Number of Sequences: 36976
Number of extensions: 121056
Number of successful extensions: 1841
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 1014
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1508
length of query: 454
length of database: 9,014,727
effective HSP length: 99
effective length of query: 355
effective length of database: 5,354,103
effective search space: 1900706565
effective search space used: 1900706565
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135505.8


