

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135504.2 - phase: 0 /pseudo
(2338 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
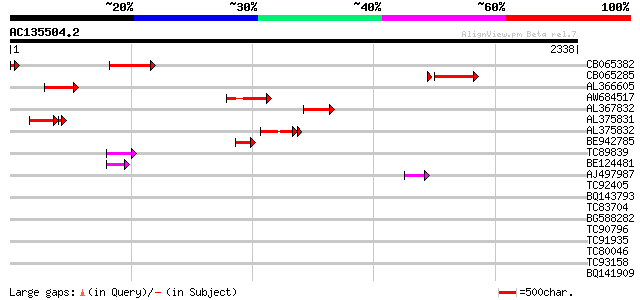
Score E
Sequences producing significant alignments: (bits) Value
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 312 1e-84
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 282 3e-79
AL366605 266 5e-71
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 263 5e-70
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 250 5e-66
AL375831 174 1e-58
AL375832 195 2e-49
BE942785 136 1e-31
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 83 1e-15
BE124481 64 5e-10
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 59 3e-08
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 37 0.090
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 35 0.45
TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19... 34 0.58
BG588282 similar to PIR|D84731|D847 probable uclacyanin I [impor... 34 0.76
TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Ar... 34 0.76
TC91935 33 0.99
TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor ... 33 0.99
TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32... 33 1.3
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 33 1.7
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 312 bits (799), Expect = 1e-84
Identities = 152/189 (80%), Positives = 161/189 (84%), Gaps = 1/189 (0%)
Frame = -2
Query: 411 QPQVPVNAIVQQMQQQPPTQQQQQ-QQARPTFPPIPMLYFELLPTLLLRGHCTTRQGKPP 469
+P +P PP QQ+Q QQARPTFP IPMLY E LPTLLLRGHCT RQGKPP
Sbjct: 569 RPMIP*LLC*PPRNNHPPVHQQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPP 390
Query: 470 PDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPN 529
PDPLPPRFRSDLKCDFHQGALGHDVEGCYALK+IVKKLI+QGKLTFENNVPHVLDNPLPN
Sbjct: 389 PDPLPPRFRSDLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPN 210
Query: 530 HVAVNMIEVCEEAPRLDVRNIATPLVPLHIKLCQASLFDHDHASCPKCFRNPLGCYVVQN 589
H AVNMIEV EEAP LDVRN+ TPLVPLHIKLCQASLFDHDHA+C + F NPLGC VVQN
Sbjct: 209 HAAVNMIEVYEEAPGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQN 30
Query: 590 DI*SLMNDN 598
DI SLMN+N
Sbjct: 29 DIQSLMNNN 3
Score = 67.8 bits (164), Expect = 5e-11
Identities = 35/40 (87%), Positives = 37/40 (92%)
Frame = -1
Query: 1 NAQLRTELASLREELAKANDTMTALLAAQEQSATVIPVTT 40
NAQLRTELAS+REELAKANDTMTALLAAQEQS+T P TT
Sbjct: 618 NAQLRTELASMREELAKANDTMTALLAAQEQSSTGSPTTT 499
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 282 bits (722), Expect(2) = 3e-79
Identities = 150/182 (82%), Positives = 156/182 (85%)
Frame = -3
Query: 1752 QIPIASGD*SSMVLSMLMVKESGQPLYPRRGITSLLPPGFCSNVQTIWPNTKRVSLGSRK 1811
QIPIA+G *S MVLSMLMVKE Q LYPRRGITSLLPPGFCSNVQTIW + K VSLGSRK
Sbjct: 551 QIPIANGV*SLMVLSMLMVKELEQSLYPRRGITSLLPPGFCSNVQTIWLSMKHVSLGSRK 372
Query: 1812 QLT*ESNTSTSMEIRHLSSTR*KANGRPTMPS*FLTVITRDVC*HILQRSNCTISLGMRT 1871
QLT*ESNTSTSMEI HLSSTR + NGRPTMP *FLT R VC* ILQ+ +CTI L RT
Sbjct: 371 QLT*ESNTSTSMEILHLSSTRSRVNGRPTMPI*FLTATMRGVC*PILQKLSCTIFLVTRT 192
Query: 1872 KWLILLLLYPPCFE*TIGMMCQ*SKCNALKDLRMCSLLGM*SIRLVKMWLTINPGITTSN 1931
KWL+L LLYPPCFE*TIGMMCQ*SKCNALKDL MC LLGM*SIR VKMWLTINPG TTSN
Sbjct: 191 KWLMLWLLYPPCFE*TIGMMCQ*SKCNALKDLHMCLLLGM*SIRPVKMWLTINPGTTTSN 12
Query: 1932 SS 1933
SS
Sbjct: 11 SS 6
Score = 33.9 bits (76), Expect(2) = 3e-79
Identities = 15/17 (88%), Positives = 17/17 (99%)
Frame = -1
Query: 1724 EIKRLRRTVDW*RSRNQ 1740
+IKRLRRTVDW*RSR+Q
Sbjct: 592 KIKRLRRTVDW*RSRSQ 542
>AL366605
Length = 422
Score = 266 bits (681), Expect = 5e-71
Identities = 123/139 (88%), Positives = 134/139 (95%)
Frame = -3
Query: 143 EIKANRGSSDTIKTHDLCLVPKVDIPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSDN 202
EIKANRG++D+ KT DLCLV KVD+PKKFK+P+FDRYNGLTCPQNHIIKYVRKMGNY DN
Sbjct: 417 EIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDN 238
Query: 203 DSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQ 262
DSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDELAAAFK+HYGFNTRLKP+REFLRSLSQ
Sbjct: 237 DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQ 58
Query: 263 KKDESFCEYAQRWRGAAAR 281
KK+ESF EYAQRWRGAAAR
Sbjct: 57 KKEESFREYAQRWRGAAAR 1
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana chromosome
II BAC F26H6; putative retroelement pol polyprotein,
partial (1%)
Length = 488
Score = 263 bits (673), Expect = 5e-70
Identities = 138/187 (73%), Positives = 145/187 (76%)
Frame = +1
Query: 892 ITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSF 951
ITFQVMDINASYSCLLGRPW HDAGAVTSTLHQKLKFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 952 SCIEAGSAEGTAFQGLTIEGTEPKKAGAAMASLKDAQKAVQEGQAVG*GKLIQLRGNKHK 1011
SAEGTAFQGL++EG EPKK GAAMASLKDAQKAVQEGQA GKLIQL NK K
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 1012 EGLGFSPTSGVSTGTFHSAGFVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVS 1071
EGL FSPTSGVSTGTFHSAGFVN + EE F PRP+FVIPGGIA+DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
Query: 1072 E*CVLLL 1078
*CVLLL
Sbjct: 466 X*CVLLL 486
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 250 bits (638), Expect = 5e-66
Identities = 122/127 (96%), Positives = 125/127 (98%)
Frame = -3
Query: 1210 RKESHSASPGRD*AYQPRYRGEQARDQGRCCSRGRG*KEDLPAPPKIPGYFCMVV*RYAR 1269
RKESHSASPGRD*AYQPRYRGEQARDQGRCCSRGRG*KEDLPAPP+IPGYFCM+V*RYAR
Sbjct: 382 RKESHSASPGRD*AYQPRYRGEQARDQGRCCSRGRG*KEDLPAPPRIPGYFCMLV*RYAR 203
Query: 1270 SRS*DCGTSDPNKA*MSSRQAEIEKDSSRYGSQD*E*GSKAD*CGFSHDSRVS*MGCQYC 1329
SRS*+CGTSDPNKA*MSSRQ EIEKDSSRYGSQD*E*GSKAD*CGFSHDS VS*MGCQYC
Sbjct: 202 SRS*NCGTSDPNKA*MSSRQVEIEKDSSRYGSQD*E*GSKAD*CGFSHDS*VS*MGCQYC 23
Query: 1330 TCAKEGW 1336
TCAKEGW
Sbjct: 22 TCAKEGW 2
>AL375831
Length = 467
Score = 174 bits (440), Expect(2) = 1e-58
Identities = 82/119 (68%), Positives = 96/119 (79%)
Frame = +2
Query: 83 IPGANLASIGITLTQATATVTEPVMITVPQGTHAGAHRGSTPITGTMEERIEELAKELRR 142
IP N ASI TL Q TA VTEP++ T+PQG + S P+T TMEE +EELAKELR
Sbjct: 8 IPATNAASIIATLPQTTAAVTEPLVHTLPQGININTQHRSIPVTKTMEEMMEELAKELRH 187
Query: 143 EIKANRGSSDTIKTHDLCLVPKVDIPKKFKVPEFDRYNGLTCPQNHIIKYVRKMGNYSD 201
EI+ANRG++D++KT DLCLV KVD+PKKFK+P+FDRYNGLTCPQNHIIKYVRKMGNY D
Sbjct: 188 EIQANRGNADSVKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 364
Score = 73.9 bits (180), Expect(2) = 1e-58
Identities = 32/34 (94%), Positives = 34/34 (99%)
Frame = +3
Query: 202 NDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDE 235
NDSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDE
Sbjct: 366 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 467
>AL375832
Length = 535
Score = 195 bits (496), Expect(2) = 2e-49
Identities = 113/167 (67%), Positives = 123/167 (72%), Gaps = 11/167 (6%)
Frame = +3
Query: 1032 FVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVSE*CVLLLKCFVFQNNNPLAL 1091
FVNAI+EEATG G RP FV PGGIA DWDAIDIPSIMHVSE*CVLLL FVF+NNNPLAL
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE*CVLLL*RFVFKNNNPLAL 182
Query: 1092 PEAKVIFSVRACLFRHRRY**KIVVSFTAFKF*CFFFP---WK*W*CQKKPKHLYFLINF 1148
P+A+VIFSVRACLFRH R ++ F CFFF + W*CQKKPK LYFLINF
Sbjct: 183 PKARVIFSVRACLFRHCR----LMNKNCRFFHDCFFFSAFFLEKW*CQKKPKRLYFLINF 350
Query: 1149 KKNLH--------KNLFLKSSNHFTQIEP**TRWAQ*PYVSSEF*IP 1187
KNLH K LFLKSSNH+ Q+EP**TR Q* Y SS+ * P
Sbjct: 351 -KNLHEKKKIQKTKKLFLKSSNHYMQVEP**TR*TQ*FYASSKL*FP 488
Score = 21.6 bits (44), Expect(2) = 2e-49
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = +2
Query: 1186 IPGV*SRR*GR**YTI*D 1203
IPG+*+ G **YT+*D
Sbjct: 482 IPGI*TEDEGG**YTV*D 535
>BE942785
Length = 460
Score = 136 bits (342), Expect = 1e-31
Identities = 67/82 (81%), Positives = 74/82 (89%)
Frame = -2
Query: 931 NGKLVTIHGEETYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTEPKKAGAAMASLKDAQKA 990
+G LVTIHGEE YL+SQLSSFSCIEAGSAEGTAFQGLT+EGTEPK+ G AMASLKDAQ+A
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 991 VQEGQAVG*GKLIQLRGNKHKE 1012
VQE QA G G+LIQLR NKHK+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 83.2 bits (204), Expect = 1e-15
Identities = 52/132 (39%), Positives = 63/132 (47%), Gaps = 7/132 (5%)
Frame = +3
Query: 399 PPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQ-------QQQQQARPTFPPIPMLYFEL 451
PP Q P QP P Q +Q P Q Q Q Q PIP+ Y +L
Sbjct: 228 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADL 407
Query: 452 LPTLLLRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQG 511
LP LL + T P+ LPP +R DL C FHQGA GHD E CY LK V+KLI+
Sbjct: 408 LPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENN 587
Query: 512 KLTFENNVPHVL 523
+F++ VL
Sbjct: 588 VWSFDDQDIKVL 623
>BE124481
Length = 538
Score = 64.3 bits (155), Expect = 5e-10
Identities = 40/102 (39%), Positives = 46/102 (44%), Gaps = 7/102 (6%)
Frame = +3
Query: 399 PPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQ-------QQQQQARPTFPPIPMLYFEL 451
PP Q P QP P Q +Q P Q Q Q Q PIP+ Y +L
Sbjct: 231 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADL 410
Query: 452 LPTLLLRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 493
LP LL + T P+ LPP +R DL C FHQGA GHD
Sbjct: 411 LPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 58.5 bits (140), Expect = 3e-08
Identities = 43/103 (41%), Positives = 61/103 (58%)
Frame = -1
Query: 1628 KDLLLSGLGRQKPASLFGESYNLVDIQNGSDKVYLRETCCYWEDCTLANALVQI*HYVQG 1687
+D+L S LG Q P SL+ S+ L I NGS++V++ E +DCT+A+A + I*+ V
Sbjct: 636 EDMLCSSLGCQAPPSLYD*SHYLAGI*NGSNQVHI*EARFDRKDCTVADATIGI*YRVPF 457
Query: 1688 SKSNQR*HSCRSSCLPTA**LPTN*VRFPR*RDHVFEIKRLRR 1730
+ N R +SC S PT * L T V *RD+V + +RL R
Sbjct: 456 PEGN*RQYSC*SLGSPTT*RLSTYQV*LS**RDYVSKDERL*R 328
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 37.0 bits (84), Expect = 0.090
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 1/119 (0%)
Frame = +2
Query: 825 KSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRKNV-LGEIDLPM 883
K+D + L D G +++ LD+L R + S F + GSR +G + LP+
Sbjct: 278 KADAAAKELEDAGGLVDLT--DNLDRLQGRWKLIYSSAFSSRTLGGSRPGPPIGRL-LPI 448
Query: 884 TIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEET 942
T+G I D + LG PW VT+TL K + V + K+ I + T
Sbjct: 449 TLGQVFQRIDILSKDFDNIVDLQLGAPWPLPPLEVTATLAHKFELVGSSKIKIIFEKTT 625
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 34.7 bits (78), Expect = 0.45
Identities = 30/96 (31%), Positives = 38/96 (39%), Gaps = 4/96 (4%)
Frame = +3
Query: 384 PPLYPYAQYSQHPFFPPFYHQY---PLPSGQPQVPVNAIVQQMQQQPPTQQQQQQQARPT 440
PPL+P + FFPPF+ P P P +P N PP P+
Sbjct: 3 PPLFPLS------FFPPFFPLLFFPPFPPPPPPIPHNI-------HPPF---------PS 116
Query: 441 FPPIPMLYFELLPTLLLRGHCTTRQG-KPPPDPLPP 475
PP P+ Y L+P + R PPP P PP
Sbjct: 117 PPPFPLPY--LIPPIFFLFLSL*RSFIPPPPSPFPP 218
>TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19.70 -
Arabidopsis thaliana, partial (15%)
Length = 716
Score = 34.3 bits (77), Expect = 0.58
Identities = 32/113 (28%), Positives = 42/113 (36%), Gaps = 9/113 (7%)
Frame = +1
Query: 373 ATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQQ 432
+TV P+ LP PY QH PP P P ++P + Q PP
Sbjct: 262 STVPPVATTSLPEPLPY----QHKEQPPIPVTLPPPPPLSKLPP---ATREQLPPPPPLS 420
Query: 433 QQQQARPTFPPIPMLYFEL---------LPTLLLRGHCTTRQGKPPPDPLPPR 476
+ A PP P +L LP L + R+ PPP PLP +
Sbjct: 421 KLPPAAREHPPPPPPSAKLPPAARERLPLPPPLSKLPPAARERLPPPSPLPEK 579
>BG588282 similar to PIR|D84731|D847 probable uclacyanin I [imported] -
Arabidopsis thaliana, partial (17%)
Length = 677
Score = 33.9 bits (76), Expect = 0.76
Identities = 25/80 (31%), Positives = 40/80 (49%), Gaps = 5/80 (6%)
Frame = +3
Query: 661 PLTSLAMVSNIAEGTTAALRSGRVCPPLFQKKAATPTTPPIDKAT--LTDVSPI---TKD 715
P+ S+ + AE T ++ S PLF+ + +PT P+ +T L SPI ++D
Sbjct: 243 PMISIIPSAAPAESTVSSAESPEASSPLFEAQVESPTLSPMIPSTEFLAPSSPIAQHSQD 422
Query: 716 VSRPSQSIEDSNLDEILRII 735
VS + S E NL + I+
Sbjct: 423 VS--ASSTEKGNLQAFISIV 476
>TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 965
Score = 33.9 bits (76), Expect = 0.76
Identities = 29/88 (32%), Positives = 35/88 (38%), Gaps = 18/88 (20%)
Frame = +2
Query: 374 TVAPINAA-------QLPPLYPYAQYSQHPFFPPF---YHQYPLPSGQPQVPVNAIVQQM 423
T AP +A+ Q+P YS P P Y+ Y P QPQ QQ
Sbjct: 701 TYAPSSASSQGGMHHQIPTPSSTESYSYAPQAYPGNSPYNSYVAPWAQPQSKSEFQTQQP 880
Query: 424 QQQ--------PPTQQQQQQQARPTFPP 443
QQ PP+ QQ Q +PT PP
Sbjct: 881 QQHMYQPQPTTPPSPQQHMYQPQPTTPP 964
>TC91935
Length = 1144
Score = 33.5 bits (75), Expect = 0.99
Identities = 33/145 (22%), Positives = 58/145 (39%), Gaps = 13/145 (8%)
Frame = +2
Query: 121 GSTPITGTMEERIEELAKELRREIKANRGSSDTIKTHDLCL-----VPKVDIPKKFKVPE 175
G+ T + + K L ++ +R SS ++ C V +V I ++ VP
Sbjct: 383 GTPHSTSVLSFTTWSIDKVLSAQLHGDRVSSIIPQSGKWCYK*RT*VSRVIIQSRYNVPN 562
Query: 176 FDRYNGLTCP--------QNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSK 227
F R LTCP N++I Y+ ++ N ++ C QD + Y L
Sbjct: 563 FSRSTALTCPTFDQEASLPNNLIAYLSRLANM----MIIFSCKQDHI-------YVRLKN 709
Query: 228 DDVHTFDELAAAFKNHYGFNTRLKP 252
+ T ++ +N +TR +P
Sbjct: 710 KGIITLAQVEGFKRN----DTRFQP 772
>TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor (EC
3.2.1.4) (Endo-1 4-beta-glucanase) (Spore germination
protein 270-6), partial (11%)
Length = 643
Score = 33.5 bits (75), Expect = 0.99
Identities = 31/102 (30%), Positives = 39/102 (37%)
Frame = +1
Query: 377 PINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQQQQQQ 436
P Q PP P + Q P P Q P P PQ P + Q++ Q+PP Q+
Sbjct: 310 PQKPPQRPPQKPPQRPPQRP--PQKLPQRP-PQKLPQRPPQKLPQKLPQKPP----QRPL 468
Query: 437 ARPTFPPIPMLYFELLPTLLLRGHCTTRQGKPPPDPLPPRFR 478
P PP+ L P L H +P P P P R
Sbjct: 469 PHPQKPPLRPLPHPQKPPLRPLPHPQKPPLRPLPHPQKPPLR 594
>TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32840
[imported] - Arabidopsis thaliana, partial (9%)
Length = 673
Score = 33.1 bits (74), Expect = 1.3
Identities = 25/78 (32%), Positives = 29/78 (37%), Gaps = 2/78 (2%)
Frame = +2
Query: 400 PFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQQQQQQARPTFPPIPMLYFELLPTLLLRG 459
PF + P P+ +P P QQQ QQQA P P LY P+
Sbjct: 86 PFTNPNPNPNSKPI-----------HYPQQQQQPQQQALHVRSPNPFLYPFASPSRASAN 232
Query: 460 HCTTRQGKPPP--DPLPP 475
H PPP P PP
Sbjct: 233 HAVGGYPPPPPPSQPQPP 286
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 32.7 bits (73), Expect = 1.7
Identities = 24/86 (27%), Positives = 38/86 (43%)
Frame = -3
Query: 372 MATVAPINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQ 431
+ ++ P+ +PP P +++ PF PP +PLP P +P ++ PP+
Sbjct: 378 LPSIFPLPPPSVPP--PLSRFPL-PFVPPPPF-FPLPLLPPPLPPPPLLPLPLSDPPSAP 211
Query: 432 QQQQQARPTFPPIPMLYFELLPTLLL 457
P FP P+L LP LL
Sbjct: 210 PPPSPPSPRFPLSPLLLILPLPLPLL 133
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.340 0.147 0.493
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,926,609
Number of Sequences: 36976
Number of extensions: 1185813
Number of successful extensions: 9833
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 5100
Number of HSP's successfully gapped in prelim test: 502
Number of HSP's that attempted gapping in prelim test: 3852
Number of HSP's gapped (non-prelim): 6544
length of query: 2338
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2226
effective length of database: 4,873,415
effective search space: 10848221790
effective search space used: 10848221790
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135504.2


