

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.6 + phase: 0
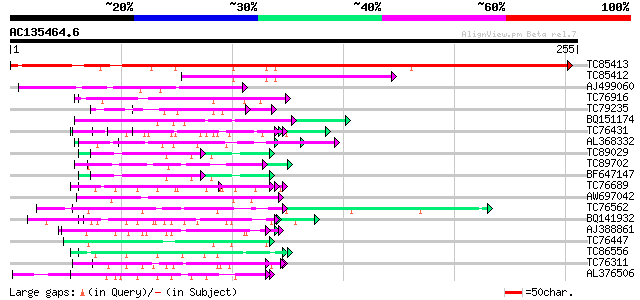
(255 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85413 similar to PIR|S23737|S23737 proline-rich protein precur... 271 2e-73
TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 108 2e-24
AJ499060 weakly similar to GP|13122418|dbj putative glycin-rich ... 69 2e-12
TC76916 MtN4 59 2e-09
TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene... 58 3e-09
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 56 2e-08
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 55 2e-08
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 55 3e-08
TC89029 weakly similar to GP|15450467|gb|AAK96527.1 At1g12810/F1... 54 6e-08
TC89702 similar to PIR|T01157|T01157 hypothetical protein F7N22.... 54 6e-08
BF647147 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophil... 54 6e-08
TC76689 MtN12 52 3e-07
AW697042 similar to GP|13122418|dbj putative glycin-rich protein... 52 3e-07
TC76562 similar to PIR|T11622|T11622 extensin class 1 precursor ... 51 5e-07
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 50 6e-07
AJ388861 weakly similar to GP|1754989|gb| proline-rich protein P... 50 6e-07
TC76447 homologue to GP|11121502|emb|CAC14888. putative extensin... 50 1e-06
TC86556 similar to PIR|T07598|T07598 proline-rich protein GPP1 -... 49 1e-06
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 49 1e-06
AL376506 homologue to GP|15021744|gb root nodule extensin {Pisum... 49 2e-06
>TC85413 similar to PIR|S23737|S23737 proline-rich protein precursor -
kidney bean, partial (56%)
Length = 1139
Score = 271 bits (693), Expect = 2e-73
Identities = 154/285 (54%), Positives = 178/285 (62%), Gaps = 32/285 (11%)
Frame = +1
Query: 1 MAKIILALIFLLLQITSFVVFAEELETLHHKPTTPLHPP--TKSPVHKPLAKPPTHAPHH 58
MAK LA++ LLLQ+ SF FAEELETL TP +PP T +P+H PP +APHH
Sbjct: 46 MAKT-LAMLLLLLQLISFTTFAEELETL-----TPTYPPHTTPAPLH-----PPANAPHH 192
Query: 59 HHHH---------------SPSHAPLPPPH-----PAKPLTHHHHQHQHHSPAPSPYHVP 98
HHHH SP H PL PP+ PAKP TH HH H H PAP+P H P
Sbjct: 193 HHHHHIHSPTPAPTPTPSPSPIHTPLHPPYHSAPVPAKPPTHGHHHHHPHPPAPTPVHTP 372
Query: 99 T-----PLQRPAKPPTLHHHQ--HPPA--HAPTHMPRVSRSSIAVEGVVYVKSCHHAGFD 149
PL P P + H HPPA H P H + RS IAV+GVVYVKSC +AG D
Sbjct: 373 VAPAHPPLHPPVHTPVVPTHPPLHPPAPAHPPLHPTPLPRSFIAVQGVVYVKSCKYAGVD 552
Query: 150 TLKGATLLFGAVVKFQCHNAKYKFVLKAKT-NKEGYIYIGSSKNISSYASGHCNVVLESA 208
TL GAT + GAVVK QC+N KYK V K +T +K GY +I KNI+SYA+ CN+VL SA
Sbjct: 553 TLLGATQILGAVVKLQCNNTKYKLVQKVQTTDKNGYFFIEGPKNITSYAAHKCNIVLISA 732
Query: 209 PNGLKPSNLHGGLTGAHPKSVKRIVSKGVSLIRYTVDPLAFEPKC 253
PNGLKPSNLHGGLTGA + K V+KG+ I YTV PLAFEPKC
Sbjct: 733 PNGLKPSNLHGGLTGAGLRPEKPFVAKGLPFILYTVGPLAFEPKC 867
>TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 3229
Score = 108 bits (270), Expect = 2e-24
Identities = 57/106 (53%), Positives = 64/106 (59%), Gaps = 9/106 (8%)
Frame = +1
Query: 78 LTHHHHQHQHHSPAPSPYHVPT-----PLQRPAKPPTLHHHQ--HPPA--HAPTHMPRVS 128
L H HH H H PAP+P H P PL P P + H HPPA H P H +
Sbjct: 2908 LPHGHHHHHPHPPAPTPVHTPVAPAHPPLHPPVHTPVVPTHPPLHPPAPAHPPLHPTPLP 3087
Query: 129 RSSIAVEGVVYVKSCHHAGFDTLKGATLLFGAVVKFQCHNAKYKFV 174
RS IAV+GVVYVKSC +AG DTL GAT + GAVVK QC+N KYK V
Sbjct: 3088 RSFIAVQGVVYVKSCKYAGVDTLLGATQILGAVVKLQCNNTKYKLV 3225
>AJ499060 weakly similar to GP|13122418|dbj putative glycin-rich protein
{Oryza sativa (japonica cultivar-group)}, partial (6%)
Length = 463
Score = 68.6 bits (166), Expect = 2e-12
Identities = 42/115 (36%), Positives = 56/115 (48%), Gaps = 12/115 (10%)
Frame = +3
Query: 5 ILALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPH-----HH 59
+L L+ LLQ++ V A E+ T HH + P HPPT +P P P ++ H +H
Sbjct: 93 VLVLVLALLQVSYLAVLAIEVSTHHH--SHPHHPPTPTPA--PAHPPSSYDIHSLSLDNH 260
Query: 60 HHHSPSH-------APLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKP 107
H+ PSH AP+ PP P+ P+ H H P P H PTP PA P
Sbjct: 261 HNKHPSHHPPTPRKAPIQPPSPS-PIPSKSLDHPHIERRPHPSHPPTPTPAPAHP 422
>TC76916 MtN4
Length = 1850
Score = 58.5 bits (140), Expect = 2e-09
Identities = 40/105 (38%), Positives = 48/105 (45%), Gaps = 8/105 (7%)
Frame = +1
Query: 30 HKPTTPLHPPT-KSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HKP P H PT K PVHKP PP +P PS P PP HP P H H + H
Sbjct: 565 HKP--PKHSPTPKPPVHKPPRYPPKPSPCP----PPSSTPKPPHHPKPPAVHPPHVPKPH 726
Query: 89 SP-APSPYHVPTPLQRP---AKPPTLH---HHQHPPAHAPTHMPR 126
P P P V P+ P KPP + + PP P ++P+
Sbjct: 727 PPYVPKPPIVKPPIVHPPYVPKPPVVKPPPYVPKPPVVRPPYVPK 861
Score = 55.1 bits (131), Expect = 3e-08
Identities = 35/95 (36%), Positives = 40/95 (41%)
Frame = +1
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA 91
P P P H P+ KPP H P P H+P P P KP + P
Sbjct: 499 PYCPYPSPKPPSHHPPIVKPPVHKP-------PKHSPTPKPPVHKP--------PRYPPK 633
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
PSP P P P KPP HH PPA P H+P+
Sbjct: 634 PSP--CPPPSSTP-KPP---HHPKPPAVHPPHVPK 720
Score = 39.7 bits (91), Expect = 0.001
Identities = 31/98 (31%), Positives = 38/98 (38%), Gaps = 2/98 (2%)
Frame = +1
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS 89
H P P P K P + P KPP P + P P+ PP+ KP
Sbjct: 772 HPPYVPKPPVVKPPPYVP--KPPVVRPPYVP--KPPVVPVTPPYVPKP------------ 903
Query: 90 PAPSPYHVPTPLQRPAKPPTLHHHQ--HPPAHAPTHMP 125
P P HVP P P+ PP + PP P H+P
Sbjct: 904 PIVFPPHVPLPPVVPSPPPYVPTPPIVKPPIVFPPHVP 1017
Score = 35.0 bits (79), Expect = 0.028
Identities = 32/94 (34%), Positives = 37/94 (39%), Gaps = 7/94 (7%)
Frame = +1
Query: 32 PTTPLHPPTKSPVH----KPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
P PL P SP P+ KPP P H PLPP P P +
Sbjct: 919 PHVPLPPVVPSPPPYVPTPPIVKPPIVFP--------PHVPLPPVVPVTP-PYVQPPPIV 1071
Query: 88 HSPAPSPYHV--PT-PLQRPAKPPTLHHHQHPPA 118
P P+P V PT P + P PP L + PPA
Sbjct: 1072 TPPTPTPPIVTPPTPPSETPCPPPPLVPYPPPPA 1173
Score = 28.5 bits (62), Expect = 2.6
Identities = 15/33 (45%), Positives = 15/33 (45%)
Frame = +2
Query: 58 HHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP 90
HH HH H L H L HHH QH H P
Sbjct: 1103 HHQHHLVKHLVL---HHHLYLIHHH-QHNKHVP 1189
Score = 26.6 bits (57), Expect = 10.0
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = +1
Query: 89 SPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSR 129
SP P +H P KPP PP H+PT P V +
Sbjct: 517 SPKPPSHHPPI-----VKPPV----HKPPKHSPTPKPPVHK 612
>TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene product
{Drosophila melanogaster}, partial (21%)
Length = 648
Score = 58.2 bits (139), Expect = 3e-09
Identities = 30/76 (39%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Frame = -2
Query: 56 PHHHHHHSPSHAP--------LPPPHPAKPLTHHHHQHQHHSP-APSP--YHVPTPLQRP 104
PH+H H+ P P L P P P+ H HH H P AP P +H P P P
Sbjct: 530 PHNHRHNPPPPGPPIMGRGPPLGPGPPPPPMPHSHHPPPPHDPFAPPPPVFHHPPPPPNP 351
Query: 105 AKPPTLHHHQHPPAHA 120
PP + HH H P HA
Sbjct: 350 YAPPPVVHHHHTPLHA 303
Score = 53.1 bits (126), Expect = 1e-07
Identities = 31/74 (41%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Frame = -2
Query: 37 HPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHP--AKPLTHHHHQHQHHSPAPSP 94
HPP P H P A PP P HH P PPP+P P+ HHHH H PAP P
Sbjct: 425 HPP---PPHDPFAPPP---PVFHH-------PPPPPNPYAPPPVVHHHHTPLHAHPAPPP 285
Query: 95 YHVPTPLQRPAKPP 108
P P +PP
Sbjct: 284 PQPGHPGPPPPRPP 243
Score = 38.5 bits (88), Expect = 0.003
Identities = 22/56 (39%), Positives = 24/56 (42%), Gaps = 7/56 (12%)
Frame = -2
Query: 29 HHKPTTPLHPPTKSP---VHKPLAKPPTHAP----HHHHHHSPSHAPLPPPHPAKP 77
HH P P H P P H P P +AP HHHH +H PPP P P
Sbjct: 428 HHPP--PPHDPFAPPPPVFHHPPPPPNPYAPPPVVHHHHTPLHAHPAPPPPQPGHP 267
Score = 34.3 bits (77), Expect = 0.048
Identities = 17/46 (36%), Positives = 20/46 (42%), Gaps = 3/46 (6%)
Frame = -2
Query: 83 HQHQHHSPAPSPYHV---PTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H H+H+ P P P + P P PP H H PP H P P
Sbjct: 527 HNHRHNPPPPGPPIMGRGPPLGPGPPPPPMPHSHHPPPPHDPFAPP 390
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 55.8 bits (133), Expect = 2e-08
Identities = 42/136 (30%), Positives = 49/136 (35%), Gaps = 12/136 (8%)
Frame = +3
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKP------------ 77
H P TP T P KPPT H SP PPP PA P
Sbjct: 6 HDPPTPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPNQ 185
Query: 78 LTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVEGV 137
T+ + + H+P +P H P P P PPT PP APT P +R +
Sbjct: 186 TTNTTNPKKQHTPT-TPTHPPRPHTPPTPPPTT--QTTPPREAPTD-PGTTRGTPPTADD 353
Query: 138 VYVKSCHHAGFDTLKG 153
HH G KG
Sbjct: 354 AQTSPAHHPGEGGGKG 401
Score = 50.1 bits (118), Expect = 8e-07
Identities = 35/102 (34%), Positives = 43/102 (41%), Gaps = 2/102 (1%)
Frame = +2
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPS-HAPLPPP-HPAKPLTHHHHQHQH 87
H P P P P H+ T PHH H P+ H P PPP HP P T H ++
Sbjct: 143 HPPPRPRPPRPPQPNHQHDKPQKTTHPHHPHTPPPTPHPPNPPPNHPNHPTTRSAHGPRN 322
Query: 88 HSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSR 129
H P +P+ P +P PP Q A AP P +R
Sbjct: 323 H-PGDAPHRRRRP-NKPGPPPRRGGGQGGRAPAPPPPPPKAR 442
Score = 39.7 bits (91), Expect = 0.001
Identities = 32/119 (26%), Positives = 36/119 (29%), Gaps = 29/119 (24%)
Frame = +3
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPT---------------------------HAPHHHHHH 62
H PTTP HPP H P PPT +P HH
Sbjct: 216 HTPTTPTHPPRP---HTPPTPPPTTQTTPPREAPTDPGTTRGTPPTADDAQTSPAHHPGE 386
Query: 63 SPSHAPLPPPHPAKPLTHHH--HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
PPP P P H HQ+Q + RP P +HPP H
Sbjct: 387 GGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEPQGKRDRPRTDP--RKKKHPPPH 557
Score = 31.2 bits (69), Expect = 0.41
Identities = 22/71 (30%), Positives = 27/71 (37%), Gaps = 5/71 (7%)
Frame = +1
Query: 29 HHKPTTPLH-PPTKSPVHKP--LAKPPTHAPHHHHHHSPSHAPLPP--PHPAKPLTHHHH 83
++ P P H PP +P P KPP H P P PP P A+P T
Sbjct: 211 NNTPPPPPHTPPDPTPPQPPPQPPKPPHHEKRPRTPEPPGGRPPPPTTPKQARPTTQERG 390
Query: 84 QHQHHSPAPSP 94
+ P P P
Sbjct: 391 GARGARPRPPP 423
Score = 30.0 bits (66), Expect = 0.90
Identities = 24/87 (27%), Positives = 32/87 (36%), Gaps = 4/87 (4%)
Frame = +1
Query: 35 PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAK---PLTHHHHQHQHHSPA 91
P PP K + PP AP H + + PP A+ P H + + +P
Sbjct: 649 PAPPPQKKQREGGKSGPP-RAPGHEEKKPAATSRKRPPRGAREKNPAAHAREREKKKTPK 825
Query: 92 PSPY-HVPTPLQRPAKPPTLHHHQHPP 117
+P H P P PP H PP
Sbjct: 826 KNPQNHNNNP---PKTPPPQKKHPPPP 897
Score = 29.3 bits (64), Expect = 1.5
Identities = 22/81 (27%), Positives = 28/81 (34%), Gaps = 6/81 (7%)
Frame = +1
Query: 47 PLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPA--PSPYHVPTPLQRP 104
P P AP + A P P A + + + P P P P P P
Sbjct: 1 PPTTPQPPAPKRPPKEKDTQARNPQPEEATKKQKRAPEKKKNPPRGQPPPPPPPPPQAPP 180
Query: 105 AKPPTLHHHQH----PPAHAP 121
KPPT ++ PP H P
Sbjct: 181 TKPPTRQTPKNNTPPPPPHTP 243
Score = 27.7 bits (60), Expect = 4.5
Identities = 24/107 (22%), Positives = 30/107 (27%), Gaps = 7/107 (6%)
Frame = +2
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHH-------HSPSHAPLPPPHPAKPLTHHHHQ 84
P P PP P P + H+ H P PPP P + H
Sbjct: 410 PAPPPPPPKAREGRTPKPDPGSKENHNRTPGKEGPATHGPPEKKTPPPTPKRRGKHTKRA 589
Query: 85 HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSS 131
P P P P PP + + AP P R +
Sbjct: 590 PGGGPPPPHPGRGGGGPGGPPPPPPKKNSERGGRAAPRGRPGTKRKN 730
Score = 27.3 bits (59), Expect = 5.9
Identities = 14/48 (29%), Positives = 18/48 (37%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAK 76
H++ P T P K P H +P P PPPHP +
Sbjct: 485 HNRTPGKEGPATHGPPEKKTPPPTPKRRGKHTKRAPGGGP-PPPHPGR 625
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 55.5 bits (132), Expect = 2e-08
Identities = 35/97 (36%), Positives = 40/97 (41%), Gaps = 13/97 (13%)
Frame = +1
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPP---PHPAKPLTHHHHQHQH---HSPA 91
PP P P P H + HHSP +P P P P P THHH+ H H HSP
Sbjct: 25 PPPYHPYPHPHPHPHPHPHPYPVHHSPPPSPKKPYKYPSPPPP-THHHYPHPHPVYHSPP 201
Query: 92 PSPYHVPTPLQRPAKPPTLHH-------HQHPPAHAP 121
P Y +P PP +HH H PP P
Sbjct: 202 PPKYKYSSP------PPPVHHVSPKPYYHSPPPPKKP 294
Score = 53.1 bits (126), Expect = 1e-07
Identities = 36/105 (34%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Frame = +1
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHH---HSPSHAPLPPPH----PAKPLTHH 81
H P H P SP KP P P HHH+ H H+P PP + P P+ H
Sbjct: 73 HPHPYPVHHSPPPSP-KKPYKYPSPPPPTHHHYPHPHPVYHSPPPPKYKYSSPPPPVHHV 249
Query: 82 HHQHQHHSPAP--SPYHV---PTPLQRPAKPPTLHHHQHPPAHAP 121
+ +HSP P PY P P+ P P ++H PP H P
Sbjct: 250 SPKPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYH--SPPVHPP 378
Score = 52.8 bits (125), Expect = 1e-07
Identities = 39/110 (35%), Positives = 43/110 (38%), Gaps = 14/110 (12%)
Frame = +2
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKP-PTHAPHHH--------HHHSPSHAPLPPPHPAKPL 78
L H T +H PT H P TH PHH HHH P+ L P +
Sbjct: 20 LLHLHITLIHTPTPIHTHTPTPTQFTTHLPHHLKNHTSIHLHHHQPTTITLT---PTQFT 190
Query: 79 THHHHQ-----HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
THHHHQ H HH S T TLHHHQ +H TH
Sbjct: 191 THHHHQNTSIPHHHHQCITSALSPTT---------TLHHHQR--SHTNTH 307
Score = 51.2 bits (121), Expect = 4e-07
Identities = 30/80 (37%), Positives = 37/80 (45%), Gaps = 3/80 (3%)
Frame = +1
Query: 45 HKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRP 104
+K + PP + P+ H H P P PHP H HHSP PSP P + P
Sbjct: 7 YKYSSPPPPYHPYPHPH------PHPHPHP-------HPYPVHHSPPPSP---KKPYKYP 138
Query: 105 AKPPTLHH---HQHPPAHAP 121
+ PP HH H HP H+P
Sbjct: 139 SPPPPTHHHYPHPHPVYHSP 198
Score = 50.8 bits (120), Expect = 5e-07
Identities = 27/70 (38%), Positives = 33/70 (46%)
Frame = +1
Query: 56 PHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
P+ + P + P P PHP P H H HHSP PSP P + P+ PP HH +
Sbjct: 4 PYKYSSPPPPYHPYPHPHP-HPHPHPHPYPVHHSPPPSP---KKPYKYPSPPPPT-HHHY 168
Query: 116 PPAHAPTHMP 125
P H H P
Sbjct: 169 PHPHPVYHSP 198
Score = 49.3 bits (116), Expect = 1e-06
Identities = 38/142 (26%), Positives = 52/142 (35%), Gaps = 25/142 (17%)
Frame = +1
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPP---------------- 71
+HH P P K P P PPTH H+ H H H+P PP
Sbjct: 91 VHHSPP----PSPKKPYKYPSPPPPTHH-HYPHPHPVYHSPPPPKYKYSSPPPPVHHVSP 255
Query: 72 -------PHPAKPLTHHHHQHQHHSP-APSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
P P KP + H P P P + P+ P P ++H PP H+ +
Sbjct: 256 KPYYHSPPPPKKPYKYSSPPPPVHPPYTPKPVYHSPPVHPPYTPHPVYHSPPPPVHSYIY 435
Query: 124 -MPRVSRSSIAVEGVVYVKSCH 144
P S V + Y+ +C+
Sbjct: 436 ASPPPPYHS*IVVDLPYLYACY 501
Score = 34.7 bits (78), Expect = 0.037
Identities = 22/79 (27%), Positives = 33/79 (40%), Gaps = 4/79 (5%)
Frame = +2
Query: 59 HHHHSPSHAPLP----PPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQ 114
H H + H P P P P + TH H ++H+ +H PT + T HHH
Sbjct: 26 HLHITLIHTPTPIHTHTPTPTQFTTHLPHHLKNHTSIHLHHHQPTTITLTPTQFTTHHH- 202
Query: 115 HPPAHAPTHMPRVSRSSIA 133
H P H + S+++
Sbjct: 203 HQNTSIPHHHHQCITSALS 259
Score = 30.0 bits (66), Expect = 0.90
Identities = 14/39 (35%), Positives = 18/39 (45%)
Frame = +1
Query: 88 HSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
+S P PYH P P P H H +P H+P P+
Sbjct: 13 YSSPPPPYH---PYPHPHPHPHPHPHPYPVHHSPPPSPK 120
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 54.7 bits (130), Expect = 3e-08
Identities = 45/133 (33%), Positives = 56/133 (41%), Gaps = 16/133 (12%)
Frame = +1
Query: 32 PTTPLHP--PTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPP------------HPAKP 77
P +P HP P SP H P PP H+ +PSH+ PPP HP P
Sbjct: 97 PPSPFHPINPPPSPFH-PFNPPPPHSIKSPPP-APSHSSPPPPPPPRKXPPPPPSHPFSP 270
Query: 78 LTHHHH--QHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIAVE 135
H+H HH APSP HV RP+ PP L PP+ +P H + IA
Sbjct: 271 PPPHNHPPPSPHHPIAPSPPHV-----RPSPPPPL-----PPSPSPYHPTVIVIVIIAFG 420
Query: 136 GVVYVKSCHHAGF 148
G+ + A F
Sbjct: 421 GIALLSMLAFALF 459
Score = 44.7 bits (104), Expect = 4e-05
Identities = 36/107 (33%), Positives = 43/107 (39%), Gaps = 3/107 (2%)
Frame = +1
Query: 30 HKPTTPLHP--PTKSPVHKPLAKPPTHAPHHHHHHSPSHA-PLPPPHPAKPLTHHHHQHQ 86
H P P HP P SP H P+ PP+ P H + P H+ PPP P+
Sbjct: 61 HLPPPPSHPLNPPPSPFH-PINPPPS--PFHPFNPPPPHSIKSPPPAPS----------- 198
Query: 87 HHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSSIA 133
H SP P P P ++ PP H PP H H P IA
Sbjct: 199 HSSPPPPP-----PPRKXPPPPPSHPFSPPPPH--NHPPPSPHHPIA 318
Score = 42.0 bits (97), Expect = 2e-04
Identities = 29/93 (31%), Positives = 33/93 (35%), Gaps = 21/93 (22%)
Frame = +1
Query: 50 KPPTHAPHHHHHHSPSHAPLPPP---HPAKPLTHHHH------QHQHHSPAPSPYH---- 96
+P + P H PSH PPP HP P H H SP P+P H
Sbjct: 34 QPNFYFPFPHLPPPPSHPLNPPPSPFHPINPPPSPFHPFNPPPPHSIKSPPPAPSHSSPP 213
Query: 97 --------VPTPLQRPAKPPTLHHHQHPPAHAP 121
P P P PP H+H P H P
Sbjct: 214 PPPPPRKXPPPPPSHPFSPPPPHNHPPPSPHHP 312
Score = 28.9 bits (63), Expect = 2.0
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 4/34 (11%)
Frame = +1
Query: 29 HHKPTTPLHPPTKSPVH----KPLAKPPTHAPHH 58
+H P +P HP SP H P PP+ +P+H
Sbjct: 283 NHPPPSPHHPIAPSPPHVRPSPPPPLPPSPSPYH 384
>TC89029 weakly similar to GP|15450467|gb|AAK96527.1 At1g12810/F13K23_4
{Arabidopsis thaliana}, partial (71%)
Length = 643
Score = 53.9 bits (128), Expect = 6e-08
Identities = 28/66 (42%), Positives = 29/66 (43%), Gaps = 14/66 (21%)
Frame = +3
Query: 37 HPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPL--------PPPHPAKPLTH------HH 82
+PP P P PP PHHHHHH P H PPP PA P H HH
Sbjct: 135 YPPPGYPGGFP---PPPPPPHHHHHHGPPHDSYQGYFDNGYPPPPPAPPQYHNYQHVDHH 305
Query: 83 HQHQHH 88
H H HH
Sbjct: 306 HHHGHH 323
Score = 45.1 bits (105), Expect = 3e-05
Identities = 29/89 (32%), Positives = 35/89 (38%), Gaps = 1/89 (1%)
Frame = +3
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS-P 90
P + PP + P PP H HH + P + PP P P HHHH H S
Sbjct: 51 PMSNYPPPGFGSPYPPPPGPPPHQ-HHEGYPPPGYPGGFPPPPPPPHHHHHHGPPHDSYQ 227
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
P P P PP H++QH H
Sbjct: 228 GYFDNGYPPP---PPAPPQYHNYQHVDHH 305
Score = 26.9 bits (58), Expect = 7.6
Identities = 16/62 (25%), Positives = 20/62 (31%)
Frame = +3
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH P H + PP P +H++ H HHHH H H
Sbjct: 189 HHHHHGPPHDSYQGYFDNGYPPPPPAPPQYHNYQHVDH-------------HHHHGH-HG 326
Query: 89 SP 90
P
Sbjct: 327 DP 332
>TC89702 similar to PIR|T01157|T01157 hypothetical protein F7N22.3 -
Arabidopsis thaliana, partial (67%)
Length = 988
Score = 53.9 bits (128), Expect = 6e-08
Identities = 37/94 (39%), Positives = 38/94 (40%), Gaps = 7/94 (7%)
Frame = +2
Query: 30 HKPTT---PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLP----PPHPAKPLTHHH 82
H PTT PL PP S H PP PH HHHH +H PLP KP HHH
Sbjct: 5 HNPTTTTTPLSPPPNSQTHH---HPPLTPPHIHHHH--THLPLPLLLRLTTATKP--HHH 163
Query: 83 HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHP 116
H H P P P K HH HP
Sbjct: 164 RPHLHGLQ-------PLPQLLPPKSHPRRHHLHP 244
Score = 46.6 bits (109), Expect = 9e-06
Identities = 30/92 (32%), Positives = 36/92 (38%)
Frame = +2
Query: 36 LHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPY 95
LH PT + PL+ PP HHH PL PPH HHHH H
Sbjct: 2 LHNPTTTTT--PLSPPPNSQTHHH-------PPLTPPH-----IHHHHTH---------- 109
Query: 96 HVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRV 127
+P PL T HH P H +P++
Sbjct: 110 -LPLPLLLRLTTATKPHHHRPHLHGLQPLPQL 202
Score = 38.5 bits (88), Expect = 0.003
Identities = 28/85 (32%), Positives = 37/85 (42%), Gaps = 11/85 (12%)
Frame = +1
Query: 58 HHHHHSPSHAPL--PPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQH 115
++HHHSP PP PA H S P P H P+P P+ PP H +Q
Sbjct: 13 NNHHHSPLSTTQFSNPPSPAA----------HSSSHPPPPHSPSP---PSPPPPHHSYQT 153
Query: 116 PP---------AHAPTHMPRVSRSS 131
PP A APT +++ S+
Sbjct: 154 PPSPTASSWTSASAPTITSQIAPSA 228
Score = 35.0 bits (79), Expect = 0.028
Identities = 25/83 (30%), Positives = 34/83 (40%), Gaps = 10/83 (12%)
Frame = +1
Query: 45 HKPLA-----KPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPY---H 96
H PL+ PP+ A H H P H+P PP P HH SP S +
Sbjct: 25 HSPLSTTQFSNPPSPAAHSSSHPPPPHSPSPPSPPPP----HHSYQTPPSPTASSWTSAS 192
Query: 97 VPTPLQR--PAKPPTLHHHQHPP 117
PT + P+ P+ + +PP
Sbjct: 193 APTITSQIAPSATPSPPYAPNPP 261
Score = 31.6 bits (70), Expect = 0.31
Identities = 24/83 (28%), Positives = 28/83 (32%), Gaps = 11/83 (13%)
Frame = +1
Query: 29 HHKP--TTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHP---------AKP 77
HH P TT P H PP H+P P H+ PP P A
Sbjct: 22 HHSPLSTTQFSNPPSPAAHSSSHPPPPHSPSPPSPPPPHHSYQTPPSPTASSWTSASAPT 201
Query: 78 LTHHHHQHQHHSPAPSPYHVPTP 100
+T S PSP + P P
Sbjct: 202 IT----SQIAPSATPSPPYAPNP 258
>BF647147 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophila
melanogaster}, partial (8%)
Length = 426
Score = 53.9 bits (128), Expect = 6e-08
Identities = 28/66 (42%), Positives = 29/66 (43%), Gaps = 14/66 (21%)
Frame = +3
Query: 37 HPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPL--------PPPHPAKPLTH------HH 82
+PP P P PP PHHHHHH P H PPP PA P H HH
Sbjct: 108 YPPPGYPGGFP---PPPPPPHHHHHHGPPHDSYQGYFDNGYPPPPPAPPQYHNYQHVDHH 278
Query: 83 HQHQHH 88
H H HH
Sbjct: 279 HHHGHH 296
Score = 45.1 bits (105), Expect = 3e-05
Identities = 29/89 (32%), Positives = 35/89 (38%), Gaps = 1/89 (1%)
Frame = +3
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS-P 90
P + PP + P PP H HH + P + PP P P HHHH H S
Sbjct: 24 PMSNYPPPGFGSPYPPPPGPPPHQ-HHEGYPPPGYPGGFPPPPPPPHHHHHHGPPHDSYQ 200
Query: 91 APSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
P P P PP H++QH H
Sbjct: 201 GYFDNGYPPP---PPAPPQYHNYQHVDHH 278
Score = 26.9 bits (58), Expect = 7.6
Identities = 16/62 (25%), Positives = 20/62 (31%)
Frame = +3
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
HH P H + PP P +H++ H HHHH H H
Sbjct: 162 HHHHHGPPHDSYQGYFDNGYPPPPPAPPQYHNYQHVDH-------------HHHHGH-HG 299
Query: 89 SP 90
P
Sbjct: 300 DP 305
>TC76689 MtN12
Length = 759
Score = 51.6 bits (122), Expect = 3e-07
Identities = 35/100 (35%), Positives = 43/100 (43%), Gaps = 8/100 (8%)
Frame = +1
Query: 28 LHHKPTTPLHPPTKSPVHK-PLAKPPTHAPHHHHHHSPS---HAPLPPPHPAKPLTHHHH 83
+HH P++ PVH P KP H+P H P H+P PP H P H
Sbjct: 91 VHHTYPKPVYHSPPPPVHTYPHPKPVYHSPPPPVHTYPKPVYHSPPPPVHTYVP----HP 258
Query: 84 QHQHHSPAPS----PYHVPTPLQRPAKPPTLHHHQHPPAH 119
+ +HSP P P HVP P+ PP H PP H
Sbjct: 259 KPVYHSPPPPVHTYPPHVPHPVYHSPPPPV---HSPPPPH 369
Score = 50.8 bits (120), Expect = 5e-07
Identities = 34/105 (32%), Positives = 45/105 (42%), Gaps = 11/105 (10%)
Frame = +1
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPS-HAPLPPPHPAKPLTHHHHQHQHHSP 90
P P+H K H P PP P HH + P H+P PP H T+ H + +HSP
Sbjct: 34 PPPPVHTYPKPVYHSP---PP---PVHHTYPKPVYHSPPPPVH-----TYPHPKPVYHSP 180
Query: 91 APSPYHVPTPLQRPAKPPT---------LHHHQHPPAHA-PTHMP 125
P + P P+ PP ++H PP H P H+P
Sbjct: 181 PPPVHTYPKPVYHSPPPPVHTYVPHPKPVYHSPPPPVHTYPPHVP 315
Score = 50.4 bits (119), Expect = 6e-07
Identities = 37/105 (35%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Frame = +1
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAP----HHHHHHSPS-HAPLPPPHP-AKPLTHH 81
+H P H P PVH KP H+P H + H P H+P PP H KP+ H
Sbjct: 46 VHTYPKPVYHSPPP-PVHHTYPKPVYHSPPPPVHTYPHPKPVYHSPPPPVHTYPKPVYHS 222
Query: 82 HHQHQH-HSPAPSP-YHVPTP---LQRPAKPPTLHHHQHPPAHAP 121
H + P P P YH P P P P ++H PP H+P
Sbjct: 223 PPPPVHTYVPHPKPVYHSPPPPVHTYPPHVPHPVYHSPPPPVHSP 357
Score = 42.4 bits (98), Expect = 2e-04
Identities = 24/79 (30%), Positives = 32/79 (40%), Gaps = 10/79 (12%)
Frame = +1
Query: 28 LHHKPTTPLH---------PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPH-PAKP 77
++H P P+H PP + P KP H+P H P H P P H P P
Sbjct: 166 VYHSPPPPVHTYPKPVYHSPPPPVHTYVPHPKPVYHSPPPPVHTYPPHVPHPVYHSPPPP 345
Query: 78 LTHHHHQHQHHSPAPSPYH 96
+ H ++ P PYH
Sbjct: 346 VHSPPPPHYYYKSPPPPYH 402
Score = 38.9 bits (89), Expect = 0.002
Identities = 26/87 (29%), Positives = 33/87 (37%), Gaps = 2/87 (2%)
Frame = +1
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHH--HHHSPSHAPLPPPHPAKPLTHHHHQHQ 86
H KP PP KP+ P H + H H+P PP H P H
Sbjct: 154 HPKPVYHSPPPPVHTYPKPVYHSPPPPVHTYVPHPKPVYHSPPPPVHTYPP---HVPHPV 324
Query: 87 HHSPAPSPYHVPTPLQRPAKPPTLHHH 113
+HSP P + P P PP +H+
Sbjct: 325 YHSPPPPVHSPPPPHYYYKSPPPPYHN 405
Score = 35.0 bits (79), Expect = 0.028
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = +1
Query: 68 PLPPPHPAKPLTHHHHQHQHHSPAPSPYHV-PTPLQRPAKPPTLHHHQHPPA--HAP 121
P PPP P H + + +HSP P +H P P+ + PP +H + HP H+P
Sbjct: 25 PSPPPPPV----HTYPKPVYHSPPPPVHHTYPKPVYH-SPPPPVHTYPHPKPVYHSP 180
Score = 30.4 bits (67), Expect = 0.69
Identities = 13/38 (34%), Positives = 19/38 (49%), Gaps = 1/38 (2%)
Frame = +1
Query: 85 HQHHSPAPSPYHV-PTPLQRPAKPPTLHHHQHPPAHAP 121
+++ SP P P H P P+ PP H + P H+P
Sbjct: 16 YKYPSPPPPPVHTYPKPVYHSPPPPVHHTYPKPVYHSP 129
Score = 26.6 bits (57), Expect = 10.0
Identities = 18/72 (25%), Positives = 27/72 (37%)
Frame = +2
Query: 60 HHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
H SP++ LP H P + H + T L P PTL +Q H
Sbjct: 2 HQRSPTNTHLPHHHQFTPTLNQSTTLLHPQFITPTLNQSTTLLHPQFTPTLTLNQSTTLH 181
Query: 120 APTHMPRVSRSS 131
P +++S+
Sbjct: 182 HPQFTLTLNQST 217
Score = 26.6 bits (57), Expect = 10.0
Identities = 24/93 (25%), Positives = 32/93 (33%), Gaps = 28/93 (30%)
Frame = +2
Query: 53 THAPHHHHHHSPSHAPLPPPHP------------------AKPLTHHHHQHQHH------ 88
TH PHHH + HP LT + HH
Sbjct: 23 THLPHHHQFTPTLNQSTTLLHPQFITPTLNQSTTLLHPQFTPTLTLNQSTTLHHPQFTLT 202
Query: 89 -SPAPSPYH---VPTPLQRPAKPPTLHHHQHPP 117
+ + +P+H PT L ++ TLHHHQ P
Sbjct: 203 LNQSTTPHHHQFTPTSLTL-SQSTTLHHHQFTP 298
>AW697042 similar to GP|13122418|dbj putative glycin-rich protein {Oryza
sativa (japonica cultivar-group)}, partial (11%)
Length = 730
Score = 51.6 bits (122), Expect = 3e-07
Identities = 33/101 (32%), Positives = 44/101 (42%), Gaps = 8/101 (7%)
Frame = -2
Query: 31 KPTTPLHPPTKSPV-HKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHS 89
+P P PP +P + PL PT + H P P PPP P P TH+ Q +
Sbjct: 717 RPPPPTPPPRPTPPPYPPLPYSPTTSSHLS---KPPPPPTPPPLPPHPPTHYSASSQCPA 547
Query: 90 PAPSPYHVPT---PLQRPAKP----PTLHHHQHPPAHAPTH 123
P+P+P P PL P P P H + P +P+H
Sbjct: 546 PSPTPPTAPAAYPPLLLPHPPHHTLPIAHFYSSPSLQSPSH 424
Score = 33.9 bits (76), Expect = 0.063
Identities = 25/89 (28%), Positives = 34/89 (38%), Gaps = 3/89 (3%)
Frame = -3
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH---HSPAPSP 94
PP +P PL P P H+P PP P L+ H H H +P P+
Sbjct: 728 PPHAAPPPPPLHLAPLPLP-------TLHSPTLPPPPLTSLSPHPHPHLPLCLLTPPPTT 570
Query: 95 YHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
P+ +RP L HP ++ H
Sbjct: 569 LLAPSAPRRPLLLLQLQLPTHPSSYHTLH 483
Score = 31.6 bits (70), Expect = 0.31
Identities = 27/80 (33%), Positives = 30/80 (36%), Gaps = 1/80 (1%)
Frame = -2
Query: 47 PLAKPPTHAPHHHHHHSPSHAPLP-PPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPA 105
P +PP P P P P PP P P T H S P P PTP P
Sbjct: 726 PARRPPPPTP------PPRPTPPPYPPLPYSPTT-----SSHLSKPPPP---PTPPPLPP 589
Query: 106 KPPTLHHHQHPPAHAPTHMP 125
PPT H+ AP+ P
Sbjct: 588 HPPT-HYSASSQCPAPSPTP 532
Score = 30.4 bits (67), Expect = 0.69
Identities = 26/91 (28%), Positives = 30/91 (32%), Gaps = 23/91 (25%)
Frame = -1
Query: 50 KPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHH-----------HQHQHHSPAPSPYHVP 98
+P T P H PS +P P P+ PL HH H S P P
Sbjct: 730 RPRTPPPPPH----PSTSPHSPSLPSTPLLSHHLLSPL*APTPTHTSPSASSPPHPLLC* 563
Query: 99 TPLQR------------PAKPPTLHHHQHPP 117
P+ R P PPT HPP
Sbjct: 562 LPVPRAVPYSSYSSSCLPTPPPTTPSTPHPP 470
Score = 30.0 bits (66), Expect = 0.90
Identities = 16/63 (25%), Positives = 24/63 (37%)
Frame = -2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHH 88
H+ ++ P+ +P P A PP PH PPH P+ H +
Sbjct: 576 HYSASSQCPAPSPTPPTAPAAYPPLLLPH-------------PPHHTLPIAHFYSSPSLQ 436
Query: 89 SPA 91
SP+
Sbjct: 435 SPS 427
>TC76562 similar to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 1391
Score = 50.8 bits (120), Expect = 5e-07
Identities = 51/194 (26%), Positives = 74/194 (37%), Gaps = 5/194 (2%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHH-HHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
++K P P P + PP+ PH +++ SP P P P P P +++
Sbjct: 17 YYKSPPPPSPSPPPPYYYKSPPPPSPIPHTPYYYKSP---PPPSPSPPPP--YYYKSPPP 181
Query: 88 HSPAP-SPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH-MPRVSRSSIAVEGVVYVKSCHH 145
SP P +PY+ +P PP +++ PP A H P + V G VY C+
Sbjct: 182 PSPIPHTPYYYKSPPPPKVLPPPYYYNSPPPPVAYPHPHPYHHALIVRVVGKVYSFRCYD 361
Query: 146 AGFDTLK-GATLLFGAVVKFQCHNAKYKFVLKAKTNKEG-YIYIGSSKNISSYASGHCNV 203
+ L GAVV+ C KT G Y + Y S C
Sbjct: 362 WEYPAKSHNKKHLQGAVVEVTCKAGSKIVKAYGKTKNNGKYAITVKDFDYVKYGSTVCKA 541
Query: 204 VLESAPNGLKPSNL 217
L + P G P N+
Sbjct: 542 ALYAPPKG-SPFNI 580
Score = 40.4 bits (93), Expect = 7e-04
Identities = 39/114 (34%), Positives = 46/114 (40%), Gaps = 1/114 (0%)
Frame = +3
Query: 13 LQITSFVVFAEELETLHHKPTT-PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPP 71
L ITS + L LHH TT PL P+H + P H H HHH+ + L P
Sbjct: 12 LTITSLLPHLHHL--LHHHTTTNPLLLHLLFPIHLTITNLPHHLHHLLHHHTTIN--LLP 179
Query: 72 PHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
H P TH HH P +H T L LH P H TH+P
Sbjct: 180 LHLLFP-THLTTTSPHHHPRFFLHHTITTL-------LLH-----PLHILTHIP 302
Score = 39.7 bits (91), Expect = 0.001
Identities = 26/84 (30%), Positives = 35/84 (40%), Gaps = 1/84 (1%)
Frame = +2
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPS-PYH 96
PP SPV+K + PP H++SP + PP P K ++ PAP Y+
Sbjct: 737 PPPPSPVYKYNSPPPPV-----HYYSPPYTYKSPPPPVKAAPTPYYYKSPPPPAPVYKYN 901
Query: 97 VPTPLQRPAKPPTLHHHQHPPAHA 120
P P PP + PP A
Sbjct: 902 SPPPPVHYYSPPYTYKSPPPPVKA 973
Score = 38.5 bits (88), Expect = 0.003
Identities = 27/87 (31%), Positives = 37/87 (42%), Gaps = 2/87 (2%)
Frame = +2
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPS-PYH 96
PP +PV+K + PP H++SP + PP P K ++ PAP Y+
Sbjct: 869 PPPPAPVYKYNSPPPPV-----HYYSPPYTYKSPPPPVKAAPTPYYYKSPPPPAPVYKYN 1033
Query: 97 VPTPLQRPAKPPTLHHHQHPPAH-APT 122
P P PP + PP APT
Sbjct: 1034SPPPPVHYYSPPYTYKSPPPPVKVAPT 1114
Score = 38.1 bits (87), Expect = 0.003
Identities = 25/84 (29%), Positives = 35/84 (40%), Gaps = 1/84 (1%)
Frame = +2
Query: 38 PPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPS-PYH 96
PP +PV+K + PP H++SP + PP P K ++ PAP Y+
Sbjct: 1001 PPPPAPVYKYNSPPPPV-----HYYSPPYTYKSPPPPVKVAPTPYYYKSPPPPAPVYKYN 1165
Query: 97 VPTPLQRPAKPPTLHHHQHPPAHA 120
P P PP + PP A
Sbjct: 1166 SPPPPVHYYSPPYTYKSPPPPVKA 1237
Score = 36.6 bits (83), Expect = 0.010
Identities = 24/102 (23%), Positives = 41/102 (39%), Gaps = 7/102 (6%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPA-------KPLTHH 81
++ P P+H + +K P AP +++ SP PPP P P+ ++
Sbjct: 764 YNSPPPPVHYYSPPYTYKSPPPPVKAAPTPYYYKSP-----PPPAPVYKYNSPPPPVHYY 928
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+ + SP P PTP + PP +++ P H
Sbjct: 929 SPPYTYKSPPPPVKAAPTPYYYKSPPPPAPVYKYNSPPPPVH 1054
Score = 36.2 bits (82), Expect = 0.013
Identities = 24/102 (23%), Positives = 41/102 (39%), Gaps = 7/102 (6%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPA-------KPLTHH 81
++ P P+H + +K P AP +++ SP PPP P P+ ++
Sbjct: 896 YNSPPPPVHYYSPPYTYKSPPPPVKAAPTPYYYKSP-----PPPAPVYKYNSPPPPVHYY 1060
Query: 82 HHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
+ + SP P PTP + PP +++ P H
Sbjct: 1061SPPYTYKSPPPPVKVAPTPYYYKSPPPPAPVYKYNSPPPPVH 1186
Score = 33.1 bits (74), Expect = 0.11
Identities = 20/81 (24%), Positives = 34/81 (41%), Gaps = 6/81 (7%)
Frame = +2
Query: 49 AKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKP- 107
AKP +A H P P P+ K ++++SP P ++ P + P
Sbjct: 650 AKPFAYASKKHFKECEKPKPSPTPYYYKSPPPPSPVYKYNSPPPPVHYYSPPYTYKSPPP 829
Query: 108 -----PTLHHHQHPPAHAPTH 123
PT ++++ PP AP +
Sbjct: 830 PVKAAPTPYYYKSPPPPAPVY 892
Score = 31.6 bits (70), Expect = 0.31
Identities = 21/89 (23%), Positives = 35/89 (38%), Gaps = 7/89 (7%)
Frame = +2
Query: 42 SPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPA-------KPLTHHHHQHQHHSPAPSP 94
S H + P +P +++ SP PPP P P+ ++ + + SP P
Sbjct: 671 SKKHFKECEKPKPSPTPYYYKSP-----PPPSPVYKYNSPPPPVHYYSPPYTYKSPPPPV 835
Query: 95 YHVPTPLQRPAKPPTLHHHQHPPAHAPTH 123
PTP + PP +++ P H
Sbjct: 836 KAAPTPYYYKSPPPPAPVYKYNSPPPPVH 922
Score = 29.3 bits (64), Expect = 1.5
Identities = 35/134 (26%), Positives = 51/134 (37%), Gaps = 26/134 (19%)
Frame = +3
Query: 30 HKPTTPLHP--------PTKSPVHKPLAKPPTHAPHHH----HHHSPSHAPLPPPHPAKP 77
H PT+ L P T + +H P T P H HHH P LP +
Sbjct: 804 HTPTSHLLPL*KRHQLLTTTNHLHHPHRFTSTTHPLHRSIIIHHHIPISHLLPL*KRHQL 983
Query: 78 LTHHHHQH-QHHSPAPSPY---------HVPT----PLQRPAKPPTLHHHQHPPAHAPTH 123
LT +H H H + +P+ H+PT PL + + T +H H P +
Sbjct: 984 LTTTNHLHLPHRFTSTTPHLHRSIIIHLHIPTSHLLPL*KWHQLLTTTNHLHHPHRFTST 1163
Query: 124 MPRVSRSSIAVEGV 137
+ RS I + +
Sbjct: 1164THHLHRSIIILHHI 1205
Score = 26.9 bits (58), Expect = 7.6
Identities = 12/41 (29%), Positives = 17/41 (41%)
Frame = +2
Query: 90 PAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRS 130
P P Y+ P P+ PP ++ PP H P +S
Sbjct: 2 PPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPIPHTPYYYKS 124
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 50.4 bits (119), Expect = 6e-07
Identities = 42/132 (31%), Positives = 54/132 (40%), Gaps = 19/132 (14%)
Frame = -1
Query: 9 IFLLLQI-TSFVVFAEELETLHHKPTTP-------LHPPTKSPVHKPLAKPPTHAPHHHH 60
+ L++Q TSF+ +HH P P L+ PT H P PP+ P H
Sbjct: 633 VLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPATLNRPTS---HTPACPPPSRPPPHPR 463
Query: 61 HHS-----PSHAPLPPPHPAKP----LTHHHHQHQHHSPAPSPYH--VPTPLQRPAKPPT 109
S P H P PP P +P L H+ Q HSP+ H +P P P PPT
Sbjct: 462 APSLSPPPPPHPPTSPPRPGRPRSSTLAPPPHR-QFHSPSALCTHCGIPPPAVPPPPPPT 286
Query: 110 LHHHQHPPAHAP 121
PP +P
Sbjct: 285 ------PPPSSP 268
Score = 46.6 bits (109), Expect = 9e-06
Identities = 39/132 (29%), Positives = 47/132 (35%), Gaps = 24/132 (18%)
Frame = -2
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAP------HHHHHHSP-----SHAPLPP--------- 71
P P+ PPT + + P +PPT P H H SP S P PP
Sbjct: 494 PPHPVRPPTHARLRSPPLRPPTLRPPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGFPP 315
Query: 72 ---PHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRV- 127
P P PL H P +P PL P PP PP H P PR
Sbjct: 314 PPSPPPPPPLPLHPPPQLSSLPPHAPSSFLLPLTSPPPPP-------PPPHPPPSPPRPH 156
Query: 128 SRSSIAVEGVVY 139
+ ++ G VY
Sbjct: 155 NHPNLGTSGWVY 120
Score = 41.2 bits (95), Expect = 4e-04
Identities = 39/109 (35%), Positives = 42/109 (37%), Gaps = 20/109 (18%)
Frame = -2
Query: 34 TPLHP----PTKSPVHKPLAKP--PTHAPHHHHHHSPSHAP--LPPPHPAKPLTHHHH-- 83
TP HP P P PL P P P H SP P L PP PA P+ H H
Sbjct: 557 TPQHPNPPPP*TDPPATPLPAPPHPVRPPTHARLRSPPLRPPTLRPPPPA-PVAHGHRPS 381
Query: 84 -QHQHHSPAPSPYHVP----TPLQRPAKPPTLHHHQHP-----PAHAPT 122
+ S P P VP P P PP L H P P HAP+
Sbjct: 380 PPRRIGSSTPPPPFVPIAGFPPPPSPPPPPPLPLHPPPQLSSLPPHAPS 234
Score = 38.1 bits (87), Expect = 0.003
Identities = 33/112 (29%), Positives = 37/112 (32%), Gaps = 24/112 (21%)
Frame = -1
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPS----HAPLPPPH-------------- 73
P P HPPT P P H HSPS H +PPP
Sbjct: 447 PPPPPHPPTSPPRPGRPRSSTLAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSP 268
Query: 74 ------PAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
P +P +Q SP+PSP P PL P P HQ H
Sbjct: 267 PALVSPPPRPFLFPPPPNQP-SPSPSP-PPPPPLPPPPPQPPQSRHQRLGIH 118
Score = 37.4 bits (85), Expect = 0.006
Identities = 32/100 (32%), Positives = 41/100 (41%), Gaps = 11/100 (11%)
Frame = -2
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAP---LPPPHPAKPLTHHH------ 82
P P PP + H L+ PT P H SPSH P P PL+ ++
Sbjct: 1127 PYHPRPPPPLATDHTLLSPRPT--PRATHPLSPSHTIPRLAPSPGLPAPLSRYYTSLSPP 954
Query: 83 --HQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHA 120
H H +P +P +P+ R PTLH PPA A
Sbjct: 953 PLHTHDTLTPPSAPAPLPSISSRRLLRPTLHLSS-PPARA 837
Score = 36.6 bits (83), Expect = 0.010
Identities = 31/99 (31%), Positives = 39/99 (39%), Gaps = 3/99 (3%)
Frame = -1
Query: 35 PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP---A 91
PLHP T+ P+ + + T H S P PPP A L H S
Sbjct: 1320 PLHPSTRRPIRRLSPRVRTRP----RHTSG*FRPRPPPARAPRLPRSTHPCSVVSSLVVV 1153
Query: 92 PSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRS 130
S H P+ A PP+ H +PP +P H R S S
Sbjct: 1152 RSTPHTLAPIS-SASPPSPRHRPYPPLTSP-HTARYSPS 1042
Score = 36.2 bits (82), Expect = 0.013
Identities = 38/142 (26%), Positives = 46/142 (31%), Gaps = 44/142 (30%)
Frame = -1
Query: 32 PTTPLHPPTKSPVHKPLA-----------KPPTHAPHHHHHHSPSHAPLPPP--HPAKPL 78
PT PPT P +PL + HA H SP + P HPA+P
Sbjct: 822 PTLRGQPPTPRPYPQPLRTRAHVGAYPRLRDAAHAVLVKAHRSPRPPHVGPSLAHPARPR 643
Query: 79 TH----------------------HHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHP 116
HHH + S +P+ + PT PA PP HP
Sbjct: 642 PGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPATLNRPTS-HTPACPPPSRPPPHP 466
Query: 117 ---------PAHAPTHMPRVSR 129
P H PT PR R
Sbjct: 465 RAPSLSPPPPPHPPTSPPRPGR 400
Score = 34.7 bits (78), Expect = 0.037
Identities = 22/76 (28%), Positives = 23/76 (29%)
Frame = -3
Query: 51 PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTL 110
P H H H H P PP P H H PA Y+ P P
Sbjct: 961 PHRHYTHTTHSHPPQPPLRSPPSPRADFYDQHSTSHRHPPALYRYYTDPPRSTP------ 800
Query: 111 HHHQHPPAHAPTHMPR 126
H PPA T R
Sbjct: 799 HSATLPPASTHTRSCR 752
Score = 33.9 bits (76), Expect = 0.063
Identities = 40/171 (23%), Positives = 50/171 (28%), Gaps = 73/171 (42%)
Frame = -1
Query: 21 FAEELETLHHKPTTPLHPPTKSPV---HKPLAKPPTHAPHHHHHHSPSHAPL-------- 69
++ L H+ P+ P ++ V H PL TH H H SP APL
Sbjct: 1053 YSPSLSLSHYPAPRPVPRPPRAAVSILHLPLPTATTHTRHTHTPLSPRSAPLHLLAPTFT 874
Query: 70 ------------------------PPPHP-AKPLTHHHH---------------QHQHHS 89
P P P +PL H H S
Sbjct: 873 TNTPPLIATRPRSTDITPTLRGQPPTPRPYPQPLRTRAHVGAYPRLRDAAHAVLVKAHRS 694
Query: 90 PAPSPYHVPTPLQRPAK----------------------PPTLHHHQHPPA 118
P P HV L PA+ PP +HHH +PPA
Sbjct: 693 PRPP--HVGPSLAHPARPRPGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPA 547
Score = 33.5 bits (75), Expect = 0.082
Identities = 34/123 (27%), Positives = 43/123 (34%), Gaps = 28/123 (22%)
Frame = -3
Query: 37 HPPTKSPVHKPLAK--PPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSPAPSP 94
HPP P P+ + PTH P H LPPP P+ P +P PS
Sbjct: 574 HPPPP*PPSIPIPRHPEPTHQP---------HPCLPPPIPSAPPPTRAFALPPSAPPPSD 422
Query: 95 Y------------------HVPTPLQ--------RPAKPPTLHHHQHPPAHAPTHMPRVS 128
VP PL+ P +PP PP H+P+ +P S
Sbjct: 421 LPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPP------PPPHSPSILPPSS 260
Query: 129 RSS 131
R S
Sbjct: 259 RLS 251
Score = 32.0 bits (71), Expect = 0.24
Identities = 32/121 (26%), Positives = 40/121 (32%), Gaps = 22/121 (18%)
Frame = -3
Query: 31 KPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHH---------------SPSHAPLP----- 70
+P P H + PL PPT H H SP P
Sbjct: 1156 RPLNPAHARSHIIRVPPLPSPPTIPSSHLAPHRALLTLSLPLTLSRASPRPPASPRRCLD 977
Query: 71 --PPHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVS 128
PP P + H+ H HS P P PL+ P P + QH +H H P +
Sbjct: 976 TTPPSPHR-----HYTHTTHSHPPQP-----PLRSPPSPRADFYDQHSTSH--RHPPALY 833
Query: 129 R 129
R
Sbjct: 832 R 830
Score = 26.9 bits (58), Expect = 7.6
Identities = 12/32 (37%), Positives = 15/32 (46%)
Frame = -2
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHH 61
H P++ L P T P P PP P H+H
Sbjct: 245 HAPSSFLLPLTSPPPPPPPPHPPPSPPRPHNH 150
>AJ388861 weakly similar to GP|1754989|gb| proline-rich protein PRP2
precursor {Lupinus luteus}, partial (24%)
Length = 491
Score = 50.4 bits (119), Expect = 6e-07
Identities = 38/117 (32%), Positives = 48/117 (40%), Gaps = 16/117 (13%)
Frame = -3
Query: 23 EELETLHHKPT-----TPLHPPTKSPVHK---PLAKPP-THAPHHHH--HHSPSHAPLPP 71
+E + LH KP + PP + P H+ P +PP H P H H +P H PP
Sbjct: 399 QEYQPLHEKPPQMKPPSEYQPPHEKPPHEHPPPEYQPPHVHPPPEHQPPHENPPHENPPP 220
Query: 72 ----PHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRP-AKPPTLHHHQHPPAHAPTH 123
PH P H++H P H P Q P KPP H PP + P H
Sbjct: 219 EYQPPHEKPP----HYEHPPPENQPPHVHPPPEYQPPHEKPP---HEHPPPEYQPPH 70
Score = 50.1 bits (118), Expect = 8e-07
Identities = 33/103 (32%), Positives = 43/103 (41%), Gaps = 9/103 (8%)
Frame = -3
Query: 24 ELETLHHKPTTPLHPPTKSPVHK---PLAKPPTHAPHHHHHHSPSHAPL---PPPHPAKP 77
E + H P PP ++P H+ P +PP P H+ H P + P PPP P
Sbjct: 300 EYQPPHVHPPPEHQPPHENPPHENPPPEYQPPHEKPPHYEHPPPENQPPHVHPPPEYQPP 121
Query: 78 LTHHHHQHQHHSPAPSPYHVPTPLQRPA---KPPTLHHHQHPP 117
H H+H P P H P + P KPP H+ PP
Sbjct: 120 --HEKPPHEHPPPEYQPPHEKPPQENPPPEYKPP----HEKPP 10
Score = 46.6 bits (109), Expect = 9e-06
Identities = 32/104 (30%), Positives = 39/104 (36%), Gaps = 6/104 (5%)
Frame = -3
Query: 24 ELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHH----HHHHSPSHAPL--PPPHPAKP 77
E + H KP PP P H + PP H P H H + P + P PPH P
Sbjct: 348 EYQPPHEKPPHEHPPPEYQPPH--VHPPPEHQPPHENPPHENPPPEYQPPHEKPPHYEHP 175
Query: 78 LTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
+ H H P P H P + P P H+ PP P
Sbjct: 174 PPENQPPHVHPPPEYQPPHEKPPHEHP-PPEYQPPHEKPPQENP 46
>TC76447 homologue to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (58%)
Length = 564
Score = 49.7 bits (117), Expect = 1e-06
Identities = 39/111 (35%), Positives = 42/111 (37%), Gaps = 16/111 (14%)
Frame = +3
Query: 25 LETLHHKPTT----PLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTH 80
L T HH T PLH T P H HHHH S +PL P TH
Sbjct: 102 LSTHHHHQFTSTSHPLHLSTHHLHQFTSTSPHLHPSTHHHHQFTSTSPL-----LHPSTH 266
Query: 81 HHHQHQHHSP--APSPYHVP--TPLQRPAKPPTLHHHQ--------HPPAH 119
H HQ SP PS +H+ T P T HHHQ HP H
Sbjct: 267 HLHQFTSTSPHLHPSTHHLHQFTSTSPHLHPSTHHHHQFTSTSHLLHPSTH 419
Score = 38.9 bits (89), Expect = 0.002
Identities = 29/87 (33%), Positives = 34/87 (38%), Gaps = 5/87 (5%)
Frame = +3
Query: 33 TTP-LHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQHHSP- 90
T+P LHP T P H HHHH S + H P THH H + SP
Sbjct: 288 TSPHLHPSTHHLHQFTSTSPHLHPSTHHHHQFTSTS-----HLLHPSTHHLHPFTNTSPH 452
Query: 91 -APSPYHVP--TPLQRPAKPPTLHHHQ 114
PS +H+P P H HQ
Sbjct: 453 LHPSTHHLPRFISTSHHLLPSIYHLHQ 533
Score = 38.5 bits (88), Expect = 0.003
Identities = 32/105 (30%), Positives = 38/105 (35%), Gaps = 15/105 (14%)
Frame = +3
Query: 30 HKPTTPLHPPTKSPVHKPLAKPPTHAPH---HHHHHSPSHAPLPPPHPAKPLTHHHHQHQ 86
H+ T+ HP H L +H H HHHH S + HP THH HQ
Sbjct: 18 HQFTSTNHPLHLFTHHLHLFTSISHHLHLSTHHHHQFTSTS-----HPLHLSTHHLHQFT 182
Query: 87 HHSPAPSP---YHVPTPLQRPAKPPTLHH---------HQHPPAH 119
SP P +H P P+ HH H HP H
Sbjct: 183 STSPHLHPSTHHHHQFTSTSPLLHPSTHHLHQFTSTSPHLHPSTH 317
Score = 35.0 bits (79), Expect = 0.028
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 3/94 (3%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+ P P++ P PV+K + PP + P + + SP PP P +
Sbjct: 185 YKSPPPPVYSPPP-PVYKYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPV---------Y 334
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
++ SP P Y P P+ + PP + PP +
Sbjct: 335 KYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPVY 436
Score = 35.0 bits (79), Expect = 0.028
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 3/94 (3%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+ P P++ P PV+K + PP + P + + SP PP P +
Sbjct: 83 YKSPPPPVYSPPP-PVYKYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPV---------Y 232
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
++ SP P Y P P+ + PP + PP +
Sbjct: 233 KYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPVY 334
Score = 35.0 bits (79), Expect = 0.028
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 3/94 (3%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+ P P++ P PV+K + PP + P + + SP PP P +
Sbjct: 236 YKSPPPPVYSPPP-PVYKYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPV---------Y 385
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
++ SP P Y P P+ + PP + PP +
Sbjct: 386 KYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPVY 487
Score = 34.3 bits (77), Expect = 0.048
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 3/94 (3%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+ P P++ P PV+K + PP + P + + SP PP P +
Sbjct: 287 YKSPPPPVYSPPP-PVYKYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPV---------Y 436
Query: 86 QHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
++ SP P Y P P+ + PP + PP +
Sbjct: 437 KYKSPPPPVYSPPPPVYKYKSPPPPVYLPPPPVY 538
Score = 31.6 bits (70), Expect = 0.31
Identities = 22/83 (26%), Positives = 35/83 (41%), Gaps = 3/83 (3%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPP---THAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQH 85
+ P P++ P PV+K + PP + P + + SP PP P +
Sbjct: 338 YKSPPPPVYSPPP-PVYKYKSPPPPVYSPPPPVYKYKSPPPPVYSPPPPV---------Y 487
Query: 86 QHHSPAPSPYHVPTPLQRPAKPP 108
++ SP P Y P P+ + PP
Sbjct: 488 KYKSPPPPVYLPPPPVYKYKSPP 556
>TC86556 similar to PIR|T07598|T07598 proline-rich protein GPP1 - potato,
partial (24%)
Length = 1250
Score = 49.3 bits (116), Expect = 1e-06
Identities = 38/119 (31%), Positives = 47/119 (38%), Gaps = 25/119 (21%)
Frame = +3
Query: 32 PTTPLH----PPTKSPVHKPLAKP--------PTHAPHHHHHHSPS----HAPLPPPHPA 75
P TP++ PP HKPL P P P +H P H PLPPP P
Sbjct: 24 PPTPVYEKPLPPPVPVYHKPLPPPTPVYEKPLPPPVPVYHKPLPPPTPVYHKPLPPPVPV 203
Query: 76 K-PLTHHHHQHQHHSPAP--------SPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
K P +H PAP P P P+++P PP + H PP P + P
Sbjct: 204 KKPCPPPKVEHPVLPPAPVYKPPPVPKPQPPPVPVKKPCPPPKVEHPVLPP--VPVYKP 374
Score = 41.2 bits (95), Expect = 4e-04
Identities = 32/109 (29%), Positives = 42/109 (38%), Gaps = 9/109 (8%)
Frame = +3
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAK-PLTHHHHQHQ 86
++HKP P P K P P + P P + P P PPP P K P +H
Sbjct: 168 VYHKPLPP-PVPVKKPCPPPKVEHPVLPPAPVYKPPPVPKPQPPPVPVKKPCPPPKVEHP 344
Query: 87 --------HHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRV 127
P P P P+++P PP + H PP P + P V
Sbjct: 345 VLPPVPVYKPPPVPKALPPPVPIKKPC-PPKIEHPVLPP--VPIYKPPV 482
Score = 36.2 bits (82), Expect = 0.013
Identities = 29/89 (32%), Positives = 35/89 (38%), Gaps = 4/89 (4%)
Frame = +3
Query: 32 PTTPLHPPT----KSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
P P++ P K PV P+ KPP P P PLP P P + HH
Sbjct: 453 PPVPIYKPPVVIPKPPV-VPIYKPPVILPPFK---KPPCPPLPTLPPLPPKSFFHHPKFG 620
Query: 88 HSPAPSPYHVPTPLQRPAKPPTLHHHQHP 116
P S +H P + P PP H HP
Sbjct: 621 KLPPKSFFHHPKYGKWPPLPPKSFFH-HP 704
Score = 32.0 bits (71), Expect = 0.24
Identities = 27/91 (29%), Positives = 33/91 (35%), Gaps = 8/91 (8%)
Frame = +3
Query: 28 LHHKPTTPLHPPTKSPVHKPLAKP----PTHAPHHHHHHSPSHAPLPPPHPAKPLTHHHH 83
++ P P P PV KP P P P + P LPPP P K
Sbjct: 258 VYKPPPVPKPQPPPVPVKKPCPPPKVEHPVLPPVPVYKPPPVPKALPPPVPIKKPCPPKI 437
Query: 84 QHQHHSPAP---SPYHVPTPLQRPA-KPPTL 110
+H P P P +P P P KPP +
Sbjct: 438 EHPVLPPVPIYKPPVVIPKPPVVPIYKPPVI 530
Score = 26.6 bits (57), Expect = 10.0
Identities = 17/56 (30%), Positives = 20/56 (35%)
Frame = +3
Query: 26 ETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLTHH 81
++ H P PP H K P P HH P + PP P K HH
Sbjct: 591 KSFFHHPKFGKLPPKSFFHHPKYGKWPPLPPKSFFHH-PKYFGKWPPIPHKSFFHH 755
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 49.3 bits (116), Expect = 1e-06
Identities = 31/104 (29%), Positives = 45/104 (42%), Gaps = 16/104 (15%)
Frame = +2
Query: 38 PPTKSP-----VHKPLAKPPTHAPHHHHHHSPSHAPL----------PPPHPAKPLTHHH 82
PP+ SP H P P+ P +++ P +P PPP P+ P +++
Sbjct: 2 PPSPSPPPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYY 181
Query: 83 HQHQHHSPAP-SPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
SP P +PYH +P A PP +H+ PP PT P
Sbjct: 182 QSPPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPP--PPTSSP 307
Score = 48.1 bits (113), Expect = 3e-06
Identities = 28/104 (26%), Positives = 45/104 (42%), Gaps = 15/104 (14%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHS-----------PSHAPLPPPHPAKP 77
++K P P P + PP+ PH +H+ P H PPP + P
Sbjct: 128 YYKSPPPPSPSPPPPYYYQSPPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTSSP 307
Query: 78 LTHHHHQHQHHSPAPSPYHV----PTPLQRPAKPPTLHHHQHPP 117
+H+ SPAP+P ++ P P++ P PP ++ + PP
Sbjct: 308 PPYHYTSPPPPSPAPAPTYIYKSPPPPMKSP--PPPVYIYASPP 433
Score = 45.8 bits (107), Expect = 2e-05
Identities = 30/109 (27%), Positives = 44/109 (39%), Gaps = 12/109 (11%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKP-PTHAPHHHHHHSPSHAPLPPP-----HPAKPLTHHH 82
+H P P P +K P P+ P +++ P +P PPP P P H
Sbjct: 35 YHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYQSPPPPSPTPH 214
Query: 83 HQHQHHSPAPS------PYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMP 125
+ + SP P PYH +P + PP +H+ PP +P P
Sbjct: 215 TPYHYKSPPPPTASPPPPYHYVSPPPPTSSPPP-YHYTSPPPPSPAPAP 358
Score = 42.7 bits (99), Expect = 1e-04
Identities = 29/100 (29%), Positives = 44/100 (44%), Gaps = 4/100 (4%)
Frame = +2
Query: 29 HHKPTTPLHPPTKSPVHKPLAKPPTHAPHH-HHHHSPSHAPLPPPHPAKPLTHHHHQHQH 87
++K P P P + PP+ +P +++ SP P P P P P H+
Sbjct: 80 YYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYQSP---PPPSPTPHTPY-HYKSPPPP 247
Query: 88 HSPAPSPYHVPTPLQRPAKPPTLHHHQHP---PAHAPTHM 124
+ P PYH +P + PP H+ P PA APT++
Sbjct: 248 TASPPPPYHYVSPPPPTSSPPPYHYTSPPPPSPAPAPTYI 367
Score = 38.9 bits (89), Expect = 0.002
Identities = 24/90 (26%), Positives = 32/90 (34%), Gaps = 2/90 (2%)
Frame = +2
Query: 32 PTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPPPHP--AKPLTHHHHQHQHHS 89
P +P PP P P H P+H+ P A PPP+ + P H++
Sbjct: 146 PPSPSPPPPYYYQSPPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTSSPPPYHYT 325
Query: 90 PAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
P P P P PP PP +
Sbjct: 326 SPPPPSPAPAPTYIYKSPPPPMKSPPPPVY 415
Score = 32.3 bits (72), Expect = 0.18
Identities = 22/67 (32%), Positives = 27/67 (39%), Gaps = 8/67 (11%)
Frame = +3
Query: 28 LHHKPTTPLHPPTKSPVHKP--------LAKPPTHAPHHHHHHSPSHAPLPPPHPAKPLT 79
LHH P L P T +P+H + P H H HH + H LPP P
Sbjct: 192 LHHLPLLIL-PTTTNPLHHQRRLLHLLTIM*VPPHLHHLHHLTTTHHHLLPPQLPLPHTY 368
Query: 80 HHHHQHQ 86
+H HQ
Sbjct: 369 TNHLPHQ 389
>AL376506 homologue to GP|15021744|gb root nodule extensin {Pisum sativum},
partial (68%)
Length = 470
Score = 48.9 bits (115), Expect = 2e-06
Identities = 43/140 (30%), Positives = 58/140 (40%), Gaps = 22/140 (15%)
Frame = +2
Query: 2 AKIILALIFLLLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPL---AKPPTHAPHH 58
A + LAL L F+ F E+ + ++P P SP P+ KP H+P
Sbjct: 29 ASLTLALAIL------FLSFPSEISANKYAYSSPPPPHYSSPPPPPVHTYPKPVYHSPPP 190
Query: 59 HHHHSPS---HAPLPP----PHPAKPLTH------HHHQHQH---HSPAPSPYHVPTP-- 100
H P H+P PP PHP KP+ H H + H H HSP P + P P
Sbjct: 191 PVHTYPKPVYHSPPPPVHKYPHP-KPVYHSPPPPVHKYPHPHPVYHSPPPPVHKYPHPHP 367
Query: 101 -LQRPAKPPTLHHHQHPPAH 119
P PP +++P H
Sbjct: 368 VYHSPPPPPPKKSYKYPSPH 427
Score = 48.1 bits (113), Expect = 3e-06
Identities = 34/102 (33%), Positives = 43/102 (41%), Gaps = 12/102 (11%)
Frame = +2
Query: 28 LHHKPTTPLHPPTKS-------PVHK-PLAKPPTHAP----HHHHHHSPSHAPLPPPHPA 75
++H P P+H K PVHK P KP H+P H + H P + PPP
Sbjct: 170 VYHSPPPPVHTYPKPVYHSPPPPVHKYPHPKPVYHSPPPPVHKYPHPHPVYHSPPPPVHK 349
Query: 76 KPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPP 117
P H H H P P P ++ K P+ HHHQ P
Sbjct: 350 YP---HPHPVYHSPPPPPP-------KKSYKYPSPHHHQFTP 445
Score = 38.5 bits (88), Expect = 0.003
Identities = 20/64 (31%), Positives = 26/64 (40%), Gaps = 4/64 (6%)
Frame = +2
Query: 64 PSHAPLPPPHPA----KPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAH 119
P H PPP P KP+ H H P P + P P+ + P ++H PP H
Sbjct: 116 PPHYSSPPPPPVHTYPKPVYHSPPPPVHTYPKPVYHSPPPPVHKYPHPKPVYHSPPPPVH 295
Query: 120 APTH 123
H
Sbjct: 296 KYPH 307
Score = 33.9 bits (76), Expect = 0.063
Identities = 20/64 (31%), Positives = 26/64 (40%), Gaps = 3/64 (4%)
Frame = +2
Query: 70 PPPH---PAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPR 126
PPPH P P H + + +HSP P + P P+ +H PP H H
Sbjct: 113 PPPHYSSPPPPPVHTYPKPVYHSPPPPVHTYPKPV---------YHSPPPPVHKYPHPKP 265
Query: 127 VSRS 130
V S
Sbjct: 266 VYHS 277
Score = 33.5 bits (75), Expect = 0.082
Identities = 30/121 (24%), Positives = 40/121 (32%)
Frame = +3
Query: 12 LLQITSFVVFAEELETLHHKPTTPLHPPTKSPVHKPLAKPPTHAPHHHHHHSPSHAPLPP 71
LL + F + + TLHH TP + H T + HH
Sbjct: 132 LLHLHRFTLTLNQSTTLHHHQFTPTLNQFTTLHHHQFTSTLTLNQYTTLHH--------- 284
Query: 72 PHPAKPLTHHHHQHQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAPTHMPRVSRSS 131
P L H HH P TP+ TL HH H + TH+P + S
Sbjct: 285 PQFTSTLIHTQSTILHH-----PQFTSTPILTQCT--TLPHHHHQRSPTNTHLPTTTSSH 443
Query: 132 I 132
+
Sbjct: 444 L 446
Score = 30.8 bits (68), Expect = 0.53
Identities = 29/99 (29%), Positives = 35/99 (35%), Gaps = 13/99 (13%)
Frame = +3
Query: 36 LHPPTKSPV-HKPLAKPPTHAPHHHHHHSP-SHAPLPPPHPAKPLTHHHHQ--------- 84
LH P+ V H + TH P HHHH + H + T HHHQ
Sbjct: 42 LH*PSSF*VSHLKFQQTNTHIPLHHHHITHLLHLHRFTLTLNQSTTLHHHQFTPTLNQFT 221
Query: 85 --HQHHSPAPSPYHVPTPLQRPAKPPTLHHHQHPPAHAP 121
H H + + T L P TL H Q H P
Sbjct: 222 TLHHHQFTSTLTLNQYTTLHHPQFTSTLIHTQSTILHHP 338
Score = 27.7 bits (60), Expect = 4.5
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 6/48 (12%)
Frame = +2
Query: 28 LHHKPTTPLHP-PTKSPV-HKPLAKPPT----HAPHHHHHHSPSHAPL 69
++H P P+H P PV H P PP + HHH +P+ L
Sbjct: 317 VYHSPPPPVHKYPHPHPVYHSPPPPPPKKSYKYPSPHHHQFTPTSLTL 460
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,610,776
Number of Sequences: 36976
Number of extensions: 464181
Number of successful extensions: 17739
Number of sequences better than 10.0: 1567
Number of HSP's better than 10.0 without gapping: 7111
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12010
length of query: 255
length of database: 9,014,727
effective HSP length: 94
effective length of query: 161
effective length of database: 5,538,983
effective search space: 891776263
effective search space used: 891776263
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135464.6


