

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.4 + phase: 0
(631 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
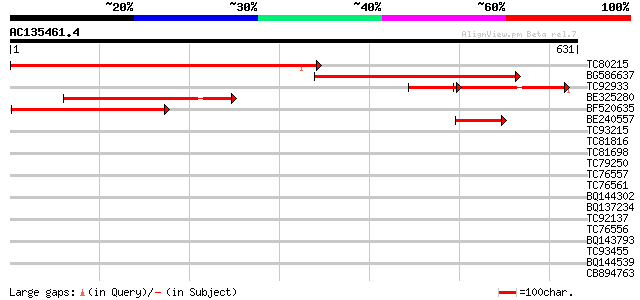
Score E
Sequences producing significant alignments: (bits) Value
TC80215 similar to GP|7547106|gb|AAF63778.1| unknown protein {Ar... 585 e-167
BG586637 similar to GP|18377985|gb unknown protein {Arabidopsis ... 471 e-133
TC92933 similar to GP|18377985|gb|AAL67135.1 unknown protein {Ar... 197 2e-73
BE325280 similar to GP|14334426|gb unknown protein {Arabidopsis ... 234 1e-61
BF520635 weakly similar to GP|11994240|db gb|AAF40458.1~gene_id:... 190 2e-48
BE240557 similar to GP|11994240|dbj gb|AAF40458.1~gene_id:MUJ8.1... 62 5e-10
TC93215 similar to GP|10177503|dbj|BAB10897. ankyrin repeat prot... 40 0.003
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 35 0.067
TC81698 similar to PIR|T09485|T09485 cold-induced protein CAP160... 35 0.11
TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 32 0.97
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 30 2.2
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 30 2.2
BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza ... 30 2.2
BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkho... 30 2.2
TC92137 similar to GP|5669650|gb|AAD46410.1| ER66 protein {Lycop... 30 2.2
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 30 2.2
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 30 2.8
TC93455 similar to PIR|D84448|D84448 probable ankyrin [imported]... 29 4.8
BQ144539 GP|13173388|g lysine/glutamic acid-rich protein {Candid... 29 6.3
CB894763 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon pi... 29 6.3
>TC80215 similar to GP|7547106|gb|AAF63778.1| unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1486
Score = 585 bits (1508), Expect = e-167
Identities = 280/358 (78%), Positives = 327/358 (91%), Gaps = 11/358 (3%)
Frame = +1
Query: 1 MEDISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVI 60
MED++KY HSPAH+AV RDHA+LRR+++ +P L K GEV E+ESL+AE++AD++SSVI
Sbjct: 412 MEDLTKYTHSPAHLAVARRDHAALRRIVTGLPQLAKAGEVKNEAESLAAELRADEVSSVI 591
Query: 61 DRRDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVI 120
DRRDVPGRETPLHL VRLRD ++AEILM+AGADWSLQNE GWS+LQEAVC REE+IA++I
Sbjct: 592 DRRDVPGRETPLHLAVRLRDHVSAEILMAAGADWSLQNEHGWSALQEAVCTREEAIAMII 771
Query: 121 ARYYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKRG 180
AR+YQPLAW+KWCRRLPR++ SA+RIRDFYMEI+FHFESSVIPFIGRIAPSDTYRIWKRG
Sbjct: 772 ARHYQPLAWAKWCRRLPRIVASATRIRDFYMEITFHFESSVIPFIGRIAPSDTYRIWKRG 951
Query: 181 ANLRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNALE 240
+NLRADMTLAGFDG RIQRSDQ+FLFLGEGY SEDGN++LP GSL+AL+HKEKE+TNALE
Sbjct: 952 SNLRADMTLAGFDGLRIQRSDQTFLFLGEGYTSEDGNLTLPHGSLLALSHKEKEVTNALE 1131
Query: 241 GAGTQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDM 300
GAGTQPTE+E+A+EVS+M QTNMYRPGIDVTQAEL+P+L+WRRQEKTE+VG+WKAKVYDM
Sbjct: 1132GAGTQPTEAEVAHEVSLMSQTNMYRPGIDVTQAELIPHLSWRRQEKTEMVGNWKAKVYDM 1311
Query: 301 LNVMVSVKSRRVPGALNDEEVFA-----------NGEDYDDVLTAEERVQLDSALRMG 347
LNVMVSVKSRRVPGA+ DEE+FA N E+YDDVLTAEER+QLDSALRMG
Sbjct: 1312LNVMVSVKSRRVPGAMTDEELFAVEDGESMINGENNEEYDDVLTAEERMQLDSALRMG 1485
>BG586637 similar to GP|18377985|gb unknown protein {Arabidopsis thaliana},
partial (27%)
Length = 690
Score = 471 bits (1211), Expect = e-133
Identities = 229/229 (100%), Positives = 229/229 (100%)
Frame = +3
Query: 340 LDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKNPNQRSS 399
LDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKNPNQRSS
Sbjct: 3 LDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKNPNQRSS 182
Query: 400 DQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFKKGVRPVLWLTQDFP 459
DQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFKKGVRPVLWLTQDFP
Sbjct: 183 DQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFKKGVRPVLWLTQDFP 362
Query: 460 LKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVTFTKFEEL 519
LKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVTFTKFEEL
Sbjct: 363 LKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVTFTKFEEL 542
Query: 520 QTPEEFSTPLSSPVSFQDAKYKESEGSTSWVSWMKGSRGMPSSDSDSHI 568
QTPEEFSTPLSSPVSFQDAKYKESEGSTSWVSWMKGSRGMPSSDSDSHI
Sbjct: 543 QTPEEFSTPLSSPVSFQDAKYKESEGSTSWVSWMKGSRGMPSSDSDSHI 689
>TC92933 similar to GP|18377985|gb|AAL67135.1 unknown protein {Arabidopsis
thaliana}, partial (33%)
Length = 698
Score = 197 bits (502), Expect(2) = 2e-73
Identities = 101/132 (76%), Positives = 109/132 (82%), Gaps = 3/132 (2%)
Frame = +1
Query: 495 FPVKVAIPIVPTIRVIVTFTKFEELQTPEEFSTPLSSPVSFQDAKYKESEGSTSWVSWMK 554
FPVKVAIPIVPTIRV+VTFTKFEELQ EEFSTPLSSP FQDAK KESEGS SW+SWMK
Sbjct: 157 FPVKVAIPIVPTIRVLVTFTKFEELQPIEEFSTPLSSPAHFQDAKSKESEGSASWISWMK 336
Query: 555 GSRGMPSSDSDSHIHSHRYKDEVDPFIIPSDYKWVDANERKRRMKAKRAKIKKNKKQTVA 614
G RG SSDSD SHRYKDE DPF IP+DYKWVDANE+KRRMKAK+AK KK+KK VA
Sbjct: 337 GGRGGQSSDSD----SHRYKDEADPFSIPADYKWVDANEKKRRMKAKKAKSKKHKKPPVA 504
Query: 615 KGRDGA---HQG 623
K DG+ H+G
Sbjct: 505 KSGDGSASQHRG 540
Score = 97.4 bits (241), Expect(2) = 2e-73
Identities = 48/58 (82%), Positives = 51/58 (87%)
Frame = +3
Query: 445 KKGVRPVLWLTQDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIP 502
+KG+RPVLWLT DFPLKTDELLPLLDILANKVKAIRRLRELLTTKLP GTF + P
Sbjct: 6 RKGLRPVLWLTPDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPHGTFSCQGCYP 179
>BE325280 similar to GP|14334426|gb unknown protein {Arabidopsis thaliana},
partial (30%)
Length = 595
Score = 234 bits (596), Expect = 1e-61
Identities = 114/193 (59%), Positives = 146/193 (75%)
Frame = +1
Query: 60 IDRRDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALV 119
+DRRDVP RETPLHL VRL D AA++L SAGAD SL N GW+ LQEA+C R IA +
Sbjct: 1 LDRRDVPLRETPLHLAVRLNDVSAAKLLASAGADVSLHNAAGWNPLQEALCRRATEIATI 180
Query: 120 IARYYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKR 179
+ R++ AW+KW RRLPR++ R+RDFYMEISFHFESSVIPF+G+IAPSDTY+IWKR
Sbjct: 181 LVRHHHRSAWAKWRRRLPRLVAVLRRMRDFYMEISFHFESSVIPFVGKIAPSDTYKIWKR 360
Query: 180 GANLRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNAL 239
NLRAD +LAGFDG +IQR+DQSFLFLG+G D ++P GSL+ L +++I +A
Sbjct: 361 DGNLRADTSLAGFDGLKIQRADQSFLFLGDG----DITHAVPSGSLLVLNRDDRKIFDAF 528
Query: 240 EGAGTQPTESEIA 252
E AG +++++A
Sbjct: 529 ENAGAPMSDADVA 567
>BF520635 weakly similar to GP|11994240|db
gb|AAF40458.1~gene_id:MUJ8.11~similar to unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 621
Score = 190 bits (482), Expect = 2e-48
Identities = 88/176 (50%), Positives = 131/176 (74%)
Frame = +1
Query: 3 DISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVIDR 62
+++ + HSP H A++ +D+A+L+ ++ +P L P E+ TE+ S++ + +A IS+V+DR
Sbjct: 94 NVAAFAHSPIHKAIILKDYANLKEILGGLPKLSNPYEIKTEAASIAEDEKAAAISAVVDR 273
Query: 63 RDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIAR 122
RDV +TPLHL+V+L D AAE+LM+AGA+ L+N +GWS+L++AV N+++ I L++ +
Sbjct: 274 RDVLNGDTPLHLSVKLGDVDAAEMLMAAGANIRLKNNEGWSALRQAVINKQDKIGLIMIK 453
Query: 123 YYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWK 178
Y KW R+ PR G+ R+RDFYMEI+FHFESSVIPFI RIAPSDTY+IWK
Sbjct: 454 YSYNEMDEKWFRKFPRYFGTMRRMRDFYMEITFHFESSVIPFISRIAPSDTYKIWK 621
>BE240557 similar to GP|11994240|dbj gb|AAF40458.1~gene_id:MUJ8.11~similar to
unknown protein {Arabidopsis thaliana}, partial (6%)
Length = 349
Score = 62.4 bits (150), Expect = 5e-10
Identities = 30/58 (51%), Positives = 44/58 (75%), Gaps = 1/58 (1%)
Frame = +1
Query: 497 VKVAIPIVPTIRVIVTFTKFEELQTPEEFSTPLSSP-VSFQDAKYKESEGSTSWVSWM 553
++VAIP+VPTIRV+VTFTKFEELQ EF+TP +SP +S +++ + ++SW W+
Sbjct: 175 IQVAIPVVPTIRVLVTFTKFEELQPLYEFATPPTSPSMSREESPATTNFSASSWFQWI 348
>TC93215 similar to GP|10177503|dbj|BAB10897. ankyrin repeat protein EMB506
{Arabidopsis thaliana}, partial (31%)
Length = 775
Score = 40.0 bits (92), Expect = 0.003
Identities = 19/53 (35%), Positives = 36/53 (67%)
Frame = +1
Query: 70 TPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIAR 122
TPLH+ V+ R+ A+IL++ GAD S +N+ G ++L ++C ++ ++ +AR
Sbjct: 124 TPLHVAVQSRNRDIAKILLANGADRSTENKDGKTALDISICYGKDFMSYDLAR 282
Score = 33.1 bits (74), Expect = 0.33
Identities = 15/51 (29%), Positives = 27/51 (52%)
Frame = +1
Query: 70 TPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVI 120
TPLH V + ++L+ D ++ + +GW+ L AV +R IA ++
Sbjct: 25 TPLHYAVEVGAKQTVKLLIKYNVDVNVADNEGWTPLHVAVQSRNRDIAKIL 177
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 35.4 bits (80), Expect = 0.067
Identities = 24/72 (33%), Positives = 41/72 (56%), Gaps = 2/72 (2%)
Frame = +3
Query: 382 KEKKGWFGWNKNPNQRSS--DQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESK 439
K++KG +K+ ++ S D++K + + +DD GE SKK K+++ K+KKK E
Sbjct: 348 KKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKK-----EED 512
Query: 440 SESEFKKGVRPV 451
+ E K VR +
Sbjct: 513 EKEEGKVSVRDI 548
>TC81698 similar to PIR|T09485|T09485 cold-induced protein CAP160 - spinach,
partial (4%)
Length = 1002
Score = 34.7 bits (78), Expect = 0.11
Identities = 37/145 (25%), Positives = 61/145 (41%), Gaps = 16/145 (11%)
Frame = +1
Query: 318 DEEVFANGEDYDDVLTAEERVQLDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLY----E 373
DE ++ E+ D L E D A ++GN + V + G+ L +
Sbjct: 199 DEIEISSAEEADKTLPPEN----DEASKLGNDEKVEHQKSENGNNVGYSLTEKLAPVYGK 366
Query: 374 NCEVNGVVKEKKGWFGWNKNPNQRSSDQ---------QKLQPEFPNDDHGEMKSK---KG 421
EV VK K +G N ++ D+ +KL+P + E+ S+ KG
Sbjct: 367 VAEVGSAVKSKV--YGTNDGTETKNGDKGVTVKDYLAEKLKPSEEDKALSEVISETLNKG 540
Query: 422 KDQNLKKKKKKGASNESKSESEFKK 446
K++ LKK+ K S KS+ F++
Sbjct: 541 KEEPLKKEDGKLDSEVEKSDKVFEE 615
>TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (34%)
Length = 1145
Score = 31.6 bits (70), Expect = 0.97
Identities = 15/52 (28%), Positives = 26/52 (49%)
Frame = +2
Query: 391 NKNPNQRSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSES 442
N+NP + L+P+ +HG+ + KG +N KK K+ K++S
Sbjct: 335 NENPESMLNIGSTLKPKGKVSNHGKSLASKGSLENQKKGPKRNIQESKKTDS 490
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 30.4 bits (67), Expect = 2.2
Identities = 22/68 (32%), Positives = 31/68 (45%)
Frame = +1
Query: 322 FANGEDYDDVLTAEERVQLDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVV 381
FAN + +D + A LD G + V Q + G+GGGG GRG G
Sbjct: 403 FANEKSMNDAIEAMNGQDLD-----GRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYG 567
Query: 382 KEKKGWFG 389
E++G+ G
Sbjct: 568 GERRGYGG 591
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 30.4 bits (67), Expect = 2.2
Identities = 22/68 (32%), Positives = 31/68 (45%)
Frame = +1
Query: 322 FANGEDYDDVLTAEERVQLDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVV 381
FAN + +D + A LD G + V Q + G+GGGG GRG G
Sbjct: 499 FANEKSMNDAIEAMNGQDLD-----GRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYG 663
Query: 382 KEKKGWFG 389
E++G+ G
Sbjct: 664 GERRGYGG 687
>BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 1047
Score = 30.4 bits (67), Expect = 2.2
Identities = 17/60 (28%), Positives = 25/60 (41%), Gaps = 1/60 (1%)
Frame = -3
Query: 385 KGWFGWN-KNPNQRSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESE 443
K W+ W K R ++K + G M GK ++ KKKK+G + K E
Sbjct: 703 KKWYEWG*KEKRIRGGGRKKWKQRDMKKKDGGMDGGNGKGRDTGKKKKRGGNGGRKKNGE 524
>BQ137234 similar to EGAD|35719|3709 hypothetical protein {Burkholderia
cepacia}, partial (6%)
Length = 1184
Score = 30.4 bits (67), Expect = 2.2
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Frame = +1
Query: 395 NQRSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKK---KKGASNESKSESE 443
N+R S +QK + E N+ + K KKG ++ KKKK ++ E K E E
Sbjct: 259 NKRRSQRQKERKE--NERKPKTKQKKGGEETRKKKKQPQRRRGEREEKGEGE 408
>TC92137 similar to GP|5669650|gb|AAD46410.1| ER66 protein {Lycopersicon
esculentum}, partial (20%)
Length = 662
Score = 30.4 bits (67), Expect = 2.2
Identities = 15/51 (29%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Frame = +3
Query: 72 LHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQ-EAVCNREESIALVIA 121
LHL L A ++++G + + ++ GW++L A C RE ++AL+++
Sbjct: 411 LHLVAALGYDWAIAPIVTSGVNINFRDVNGWTALHWAASCGRERTVALLVS 563
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 30.4 bits (67), Expect = 2.2
Identities = 22/68 (32%), Positives = 31/68 (45%)
Frame = +2
Query: 322 FANGEDYDDVLTAEERVQLDSALRMGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVV 381
FAN + +D + A LD G + V Q + G+GGGG GRG G
Sbjct: 245 FANEKSMNDAIEAMNGQDLD-----GRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYG 409
Query: 382 KEKKGWFG 389
E++G+ G
Sbjct: 410 GERRGYGG 433
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 30.0 bits (66), Expect = 2.8
Identities = 25/88 (28%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Frame = -2
Query: 361 GGGGFD-GRGNLYENCEVN----GVVKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGE 415
G G+D G+G+ + E G V ++KG W + + ++K GE
Sbjct: 625 GKRGWDRGKGSKGQETENGRRK*GKVGKRKGESKWEQGQGEGGDKRKK--------GRGE 470
Query: 416 MKSKKGKDQNLKKKKKKGASNESKSESE 443
K KGK Q K+ KKG + E E
Sbjct: 469 GKGGKGKTQRGKRGGKKGGRRRGRRERE 386
>TC93455 similar to PIR|D84448|D84448 probable ankyrin [imported] -
Arabidopsis thaliana, partial (51%)
Length = 471
Score = 29.3 bits (64), Expect = 4.8
Identities = 19/70 (27%), Positives = 33/70 (47%)
Frame = +2
Query: 51 IQADQISSVIDRRDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVC 110
+ D + VI+ D G PLH + + E L++ GAD +++N G ++L A
Sbjct: 239 LSCDASAEVINSGDEEGW-APLHSAASIGNSEILEALLNKGADVNIKNNGGRAALHYAAS 415
Query: 111 NREESIALVI 120
IA ++
Sbjct: 416 KGRMKIAEIL 445
>BQ144539 GP|13173388|g lysine/glutamic acid-rich protein {Candida albicans},
partial (1%)
Length = 476
Score = 28.9 bits (63), Expect = 6.3
Identities = 25/86 (29%), Positives = 37/86 (42%), Gaps = 5/86 (5%)
Frame = +1
Query: 368 RGNLYENCEVNGVVKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGE-----MKSKKGK 422
RGN G EK+G G K +R + ++ + E P E KSK+ K
Sbjct: 4 RGNSGNKQGKGGS*NEKEGRGGSLKGRKRRKPEMKRKEEEEPERKEKEEAEILKKSKEKK 183
Query: 423 DQNLKKKKKKGASNESKSESEFKKGV 448
+ ++KKK K E+K + K V
Sbjct: 184 EAKIQKKKMK--KKEAKIRKQAAKKV 255
>CB894763 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon
pimpinellifolium}, partial (2%)
Length = 813
Score = 28.9 bits (63), Expect = 6.3
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Frame = +1
Query: 117 ALVIARYYQPLAWSKWCRRLPRVIGSA-SRIRDFYME 152
AL I R AW KWC V A S +R+FY+E
Sbjct: 97 ALEILRIESMSAWEKWCFDAENVGSRAFSHLREFYIE 207
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,932,652
Number of Sequences: 36976
Number of extensions: 244639
Number of successful extensions: 1357
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1292
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1341
length of query: 631
length of database: 9,014,727
effective HSP length: 102
effective length of query: 529
effective length of database: 5,243,175
effective search space: 2773639575
effective search space used: 2773639575
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135461.4


