

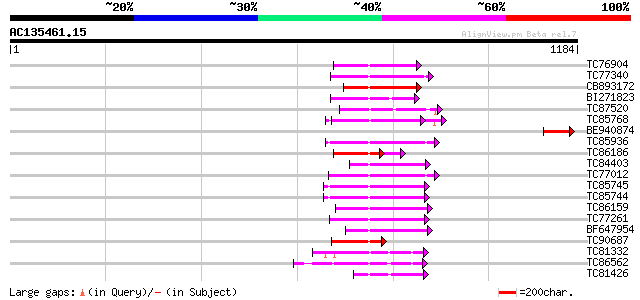
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.15 + phase: 0 /pseudo
(1184 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein... 145 1e-34
TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog ... 144 3e-34
CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like... 139 5e-33
BI271823 similar to GP|21592745|gb cell division protein FtsH-li... 136 4e-32
TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division prote... 135 7e-32
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 121 2e-27
BE940874 weakly similar to GP|6721152|gb| unknown protein {Arabi... 119 9e-27
TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidops... 117 2e-26
TC86186 similar to GP|4325041|gb|AAD17230.1| FtsH-like protein P... 94 4e-25
TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotea... 112 1e-24
TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulator... 110 3e-24
TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome re... 105 8e-23
TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15... 105 1e-22
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 104 2e-22
TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AA... 102 7e-22
BF647954 homologue to GP|11094192|db 26S proteasome regulatory p... 99 7e-21
TC90687 similar to GP|9759220|dbj|BAB09632.1 cell division prote... 99 1e-20
TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 93 5e-19
TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regu... 91 3e-18
TC81426 similar to GP|6016719|gb|AAF01545.1| putative cell divis... 90 6e-18
>TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein Pftf
precursor {Nicotiana tabacum}, partial (91%)
Length = 2667
Score = 145 bits (365), Expect = 1e-34
Identities = 80/184 (43%), Positives = 108/184 (58%)
Frame = +1
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + +D K++ EVV FL+ P F +GAR P+GVL+VG GTGKT LA AIA E
Sbjct: 820 VTFDDVAGVDEAKQDFMEVVEFLKKPERFTSVGARIPKGVLLVGPPGTGKTLLAKAIAGE 999
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP I + M+VG AS VR+LF+ A++ AP I+FV++ D RG I
Sbjct: 1000 AGVPFFSISGSEF-VEMFVGIGASRVRDLFKKAKENAPCIVFVDEIDAVGRQRGTGIGGG 1176
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQLL E+DGFE GV+++A T +D AL RPGR DR + P R
Sbjct: 1177 NDEREQTLNQLLTEMDGFEGNTGVIVVAATNRADILDSALLRPGRFDRQVSVDVPDVRGR 1356
Query: 857 ENIL 860
IL
Sbjct: 1357 TEIL 1368
>TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog chloroplast
precursor (EC 3.4.24.-). [Alfalfa] {Medicago sativa},
complete
Length = 2473
Score = 144 bits (362), Expect = 3e-34
Identities = 83/216 (38%), Positives = 120/216 (55%)
Frame = +3
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
+ V + + + + D K E+ EVV FL+NP + +GA+ P+G L+VG GTGKT L
Sbjct: 978 QEVPETGVTFADVAGADQAKLELQEVVDFLKNPDKYTALGAKIPKGCLLVGPPGTGKTLL 1157
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR 789
A A+A EA P A + ++VG AS VR+LF+ A+ AP I+F+++ D R
Sbjct: 1158 ARAVAGEAGTPFFSCAASEF-VELFVGVGASRVRDLFEKAKSKAPCIVFIDEIDAVGRQR 1334
Query: 790 GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
G + N + E INQLL E+DGF GV+++A T +D AL RPGR DR +
Sbjct: 1335 GAGLGGGNDEREQTINQLLTEMDGFSGNSGVIVLAATNRPDVLDSALLRPGRFDRQVTVD 1514
Query: 850 RPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
RP A R IL ++ L + VD+ K+A +T
Sbjct: 1515 RPDVAGRVKILQVHSR---GKALAKDVDFDKIARRT 1613
>CB893172 homologue to PIR|T45642|T45 FtsH metalloproteinase-like protein -
Arabidopsis thaliana, partial (25%)
Length = 661
Score = 139 bits (351), Expect = 5e-33
Identities = 75/164 (45%), Positives = 103/164 (62%), Gaps = 1/164 (0%)
Frame = +3
Query: 698 FLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQ 757
FL+NP + +GAR PRGVL+VG GTGKT LA A+A EA VP + A + ++VG
Sbjct: 39 FLRNPDRYARLGARPPRGVLLVGLPGTGKTLLAKAVAGEADVPFISCSASEF-VELYVGM 215
Query: 758 SASNVRELFQTARDLAPVILFVEDFDLFAGVR-GKFIHTENQDHEAFINQLLVELDGFEK 816
AS VR+LF A+ AP I+F+++ D A R GKF N + E +NQLL E+DGF+
Sbjct: 216 GASRVRDLFARAKKEAPSIIFIDEIDAVAKSRDGKFRIVGNDEREQTLNQLLTEMDGFDS 395
Query: 817 QDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENIL 860
V+++A T +D AL+RPGR DRI ++ P + RE+IL
Sbjct: 396 NSPVIVLAATNRADVLDPALRRPGRFDRIVMVETPDRIGRESIL 527
>BI271823 similar to GP|21592745|gb cell division protein FtsH-like protein
{Arabidopsis thaliana}, partial (33%)
Length = 714
Score = 136 bits (343), Expect = 4e-32
Identities = 73/187 (39%), Positives = 111/187 (59%)
Frame = +3
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
++ K + + +DS K E+ E+V+ LQ +Q++GA+ PRGVL+VG GTGKT L
Sbjct: 69 RKPKSQTVGFEDVQGVDSAKVELMEIVSCLQGDINYQKLGAKLPRGVLLVGPPGTGKTLL 248
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR 789
A A+A EA VP + A + M+VG+ A+ +R+LF AR AP I+F+++ D G R
Sbjct: 249 ARAVAGEAGVPFFTVSASEF-VEMFVGRGAARIRDLFSRARKFAPSIIFIDELDAVGGKR 425
Query: 790 GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
G+ N++ + +NQLL E+DGFE + VV++A T + +D AL RPGR +
Sbjct: 426 GRGF---NEERDQTLNQLLTEMDGFESEIRVVVIAATNRPEALDPALCRPGRFSXKVFVG 596
Query: 850 RPTQAER 856
P + R
Sbjct: 597 EPDEEGR 617
>TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division protein FtsH
protease-like {Arabidopsis thaliana}, partial (53%)
Length = 1667
Score = 135 bits (341), Expect = 7e-32
Identities = 88/224 (39%), Positives = 122/224 (54%), Gaps = 9/224 (4%)
Frame = +2
Query: 689 KEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQ 748
K+E+ EVV +L+NP F +G + P+G+L+ G GTGKT LA AIA EA VP +
Sbjct: 5 KQELEEVVEYLKNPAKFTRLGGKLPKGILLTGAPGTGKTLLAKAIAGEAGVPFFYRAGSE 184
Query: 749 LEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI-HTENQDHEAFINQL 807
E M+VG A VR LFQ A+ AP I+F+++ D R ++ HT+ H QL
Sbjct: 185 FEE-MFVGVGARRVRSLFQAAKKKAPCIIFIDEIDAVGSTRKQWEGHTKKTLH-----QL 346
Query: 808 LVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENI--LYSAAK 865
LVE+DGFE+ +G++LMA T +D AL RPGR DR + P R+ I LY K
Sbjct: 347 LVEMDGFEQNEGIILMAATNLPDILDPALTRPGRFDRHIVVPNPDVRGRQEILELYLHDK 526
Query: 866 ETMDDQLVEYVDWKKVAEKTA------LLRPIELKLVPIALEGS 903
T D+ VD K +A T L + + + A+EG+
Sbjct: 527 PTADN-----VDIKAIARGTPGFNGADLANLVNIAAIKAAVEGA 643
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 121 bits (303), Expect = 2e-27
Identities = 66/197 (33%), Positives = 107/197 (53%), Gaps = 1/197 (0%)
Frame = +2
Query: 672 VKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLA 730
V+ P ++ ++++K E+ E V + +++P F++ G +GVL G G GKT LA
Sbjct: 1511 VEVPNCSWDDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLA 1690
Query: 731 MAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRG 790
AIA E + + IK +L MW G+S +NVRE+F AR AP +LF ++ D A RG
Sbjct: 1691 KAIANECQANFISIKGPELLT-MWFGESEANVREIFDKARGSAPCVLFFDELDSIATQRG 1867
Query: 791 KFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQR 850
+ + +NQLL E+DG + V ++ T ID AL RPGR+D++ ++
Sbjct: 1868 SSVGDAGGAADRVLNQLLTEMDGMSAKKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPL 2047
Query: 851 PTQAERENILYSAAKET 867
P + R I + +++
Sbjct: 2048 PDEDSRHQIFKACLRKS 2098
Score = 101 bits (251), Expect = 2e-21
Identities = 75/268 (27%), Positives = 134/268 (49%), Gaps = 15/268 (5%)
Frame = +2
Query: 660 DPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLI 718
+PIK E+ + + ++ + +I E+V L++P+ F+ +G + P+G+L+
Sbjct: 668 EPIKREDEN----RLDEVGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILL 835
Query: 719 VGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILF 778
G G+GKT +A A+A E I ++ + + G+S SN+R+ F+ A AP I+F
Sbjct: 836 YGPPGSGKTLIARAVANETGAFFFCINGPEIMSKL-AGESESNLRKAFEEAEKNAPSIIF 1012
Query: 779 VEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQR 838
+++ D A R K T + ++QLL +DG + + V++M T ID AL+R
Sbjct: 1013IDEIDSIAPKREK---THGEVERRIVSQLLTLMDGLKSRAHVIVMGATNRPNSIDPALRR 1183
Query: 839 PGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT------------- 885
GR DR + P + R +L T + +L E VD +K++++T
Sbjct: 1184FGRFDREIDIGVPDEVGRLEVL---RIHTKNMKLAEDVDLEKISKETHGYVGADLAALCT 1354
Query: 886 -ALLRPIELKLVPIALEGSAFRSKVLDT 912
A L+ I K+ I LE +++L++
Sbjct: 1355EAALQCIREKMDVIDLEDETIDAEILNS 1438
>BE940874 weakly similar to GP|6721152|gb| unknown protein {Arabidopsis
thaliana}, partial (5%)
Length = 567
Score = 119 bits (297), Expect = 9e-27
Identities = 62/63 (98%), Positives = 62/63 (98%)
Frame = +1
Query: 1116 LLL**CGCYIKHGR*P*VCDDS*SSKDV*FGISQSKRNAAEKSPGTRKDCRGIT*I*NFD 1175
LLL* CGCYIKHGR*P*VCDDS*SSKDV*FGISQSKRNAAEKSPGTRKDCRGIT*I*NFD
Sbjct: 1 LLL*QCGCYIKHGR*P*VCDDS*SSKDV*FGISQSKRNAAEKSPGTRKDCRGIT*I*NFD 180
Query: 1176 *KG 1178
*KG
Sbjct: 181 *KG 189
>TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidopsis
thaliana}, partial (98%)
Length = 1633
Score = 117 bits (294), Expect = 2e-26
Identities = 79/241 (32%), Positives = 124/241 (50%), Gaps = 3/241 (1%)
Frame = +1
Query: 659 VDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVL 717
VDP+ MK K P + +D +EI EV+ +++P F+ +G P+GVL
Sbjct: 568 VDPLVNL---MKVEKVPDSTYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQPKGVL 738
Query: 718 IVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVIL 777
+ G GTGKT LA A+A + + +L ++G+ + VRELF AR+ AP I+
Sbjct: 739 LYGPPGTGKTLLARAVAHHTDCTFIRVSGSEL-VQKYIGEGSRMVRELFVMAREHAPSII 915
Query: 778 FVEDFDLFAGVRGKFIHTENQDHEA--FINQLLVELDGFEKQDGVVLMATTRNLKQIDEA 835
F+++ D R + + N D E + +LL +LDGFE + + ++ T + +D+A
Sbjct: 916 FMDEIDSIGSARMES-GSGNGDSEVQRTMLELLNQLDGFEASNKIKVLMATNRIDILDQA 1092
Query: 836 LQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKL 895
L RPGR+DR P + R +IL ++ L+ +D KK+AEK ELK
Sbjct: 1093LLRPGRIDRKIEFPNPNEESRRDILKIHSRRM---NLMRGIDLKKIAEKMNGASGAELKA 1263
Query: 896 V 896
V
Sbjct: 1264V 1266
>TC86186 similar to GP|4325041|gb|AAD17230.1| FtsH-like protein Pftf
precursor {Nicotiana tabacum}, partial (47%)
Length = 1303
Score = 94.4 bits (233), Expect(2) = 4e-25
Identities = 48/107 (44%), Positives = 69/107 (63%)
Frame = +2
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + +D K++ EVV FL+ P F +GAR P+GVL++G GTGKT LA AIA E
Sbjct: 845 VTFDDVAGVDEAKQDFMEVVEFLKKPERFTAIGARIPKGVLLIGPPGTGKTLLAKAIAGE 1024
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFD 783
A VP I + M+VG AS VR+LF+ A++ AP I+FV++ D
Sbjct: 1025AGVPFFSISGSEF-VEMFVGIGASRVRDLFKKAKENAPCIVFVDEID 1162
Score = 40.0 bits (92), Expect(2) = 4e-25
Identities = 20/52 (38%), Positives = 30/52 (57%)
Frame = +3
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATT 826
V+ + L+ G RG I N + E +NQLL E+DGFE G++++A T
Sbjct: 1137 VLFLLMKLMLWEGNRGTGIGGGNDEREHTLNQLLTEMDGFEGNTGIIVIAPT 1292
>TC84403 homologue to GP|15021761|gb|AAK77908.1 AAA-metalloprotease FtsH
{Pisum sativum}, partial (36%)
Length = 897
Score = 112 bits (279), Expect = 1e-24
Identities = 68/172 (39%), Positives = 93/172 (53%), Gaps = 1/172 (0%)
Frame = +3
Query: 709 GARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQT 768
GA+ P+G L+VG GTGKT LA A A E+ VP + I ++VG +S VR LF+
Sbjct: 3 GAKIPKGALLVGPPGTGKTLLAKATAGESGVPFLSISGSDF-LELFVGVGSSRVRNLFKE 179
Query: 769 ARDLAPVILFVEDFDLFAGVRG-KFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTR 827
AR AP I+F+++ D RG + N + E+ +NQLLVE+DGF GVV++A T
Sbjct: 180 ARKCAPSIVFIDEIDAIGRARGSRGGDRANDERESTLNQLLVEMDGFGTTAGVVVLAGTN 359
Query: 828 NLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWK 879
+D+AL RPGR DR + +P R+ I K D Y K
Sbjct: 360 RPDVLDKALLRPGRFDRQITIDKPDIKGRDQIFQIYLKGIKLDHEPSYYSHK 515
>TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+), complete
Length = 1793
Score = 110 bits (275), Expect = 3e-24
Identities = 71/237 (29%), Positives = 125/237 (51%), Gaps = 7/237 (2%)
Frame = +1
Query: 667 EHMKRVKKPPI---PLNNFSSIDSMKEEISEVVAFLQNPRA----FQEMGARAPRGVLIV 719
E+ RVK + P +++ I ++++I E+V + P FQ++G R P+GVL+
Sbjct: 583 EYDSRVKAMEVDEKPTEDYNDIGGLEKQIQELVEAIVLPMTHKERFQKLGIRPPKGVLLY 762
Query: 720 GERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFV 779
G GTGKT +A A AA+ +++ QL M++G A VR+ FQ A++ +P I+F+
Sbjct: 763 GPPGTGKTLMARACAAQTNATFLKLAGPQL-VQMFIGDGAKLVRDAFQLAKEKSPCIIFI 939
Query: 780 EDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRP 839
++ D R + +++ + + +LL +LDGF D + ++A T +D AL R
Sbjct: 940 DEIDAIGTKRFDSEVSGDREVQRTMLELLNQLDGFSSDDRIKVIAATNRADILDPALMRS 1119
Query: 840 GRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKLV 896
GR+DR PT+ R IL +++ + V+++++A T +LK V
Sbjct: 1120GRLDRKIEFPHPTEEARARILQIHSRKM---NVHPDVNFEELARSTDDFNGAQLKAV 1281
>TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome regulatory
particle triple-A ATPase subunit2b {Oryza sativa
(japonica cultivar-group), partial (98%)
Length = 1696
Score = 105 bits (263), Expect = 8e-23
Identities = 69/225 (30%), Positives = 117/225 (51%), Gaps = 4/225 (1%)
Frame = +3
Query: 656 REGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPR 714
++ VDP+ + MK K P + +D+ +EI E V L +P ++++G + P+
Sbjct: 588 QDEVDPMVSV---MKVEKAPLESYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPK 758
Query: 715 GVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAP 774
GV++ GE GTGKT LA A+A + + +L ++G VRELF+ A DL+P
Sbjct: 759 GVILYGEPGTGKTLLAKAVANSTSATFLRVVGSEL-IQKYLGDGPKLVRELFRVADDLSP 935
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDE 834
I+F+++ D R ++ + + +LL +LDGF+ + V ++ T ++ +D
Sbjct: 936 AIVFIDEIDAVGTKRYDAHSGGEREIQRTMLELLNQLDGFDSRGDVKVILATNRIESLDP 1115
Query: 835 ALQRPGRMDRIFHLQRPTQAERENI--LYSAAKETMDD-QLVEYV 876
AL RPGR+DR P R I ++++ DD L E+V
Sbjct: 1116ALLRPGRIDRKIEFPLPDIKTRRRIFQIHTSRMTLADDVNLEEFV 1250
>TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15.70 -
Arabidopsis thaliana, complete
Length = 1629
Score = 105 bits (261), Expect = 1e-22
Identities = 69/225 (30%), Positives = 117/225 (51%), Gaps = 4/225 (1%)
Frame = +2
Query: 656 REGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPR 714
++ VDP+ + MK K P + +D+ +EI E V L +P ++++G + P+
Sbjct: 590 QDEVDPMVSV---MKVEKAPLESYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPK 760
Query: 715 GVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAP 774
GV++ GE GTGKT LA A+A + + +L ++G VRELF+ A DL+P
Sbjct: 761 GVILYGEPGTGKTLLAKAVANSTSATFLRVVGSEL-IQKYLGDGPKLVRELFRVADDLSP 937
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDE 834
I+F+++ D R ++ + + +LL +LDGF+ + V ++ T ++ +D
Sbjct: 938 SIVFIDEIDAVGTKRYDAHSGGEREIQRTMLELLNQLDGFDSRGDVKVILATNRIESLDP 1117
Query: 835 ALQRPGRMDRIFHLQRPTQAERENI--LYSAAKETMDD-QLVEYV 876
AL RPGR+DR P R I ++++ DD L E+V
Sbjct: 1118ALLRPGRIDRKIEFPLPDIKTRRRIFTIHTSRMTLADDVNLEEFV 1252
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 104 bits (259), Expect = 2e-22
Identities = 64/207 (30%), Positives = 110/207 (52%), Gaps = 4/207 (1%)
Frame = +2
Query: 681 NFSSIDSMKEEISE----VVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
++S++ + ++I E + L NP F +G + P+GVL+ G GTGKT LA AIA+
Sbjct: 479 SYSAVGGLSDQIRELRESIELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASN 658
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
+++ + + ++G+SA +RE+F ARD P I+F+++ D G R +
Sbjct: 659 IDANFLKVVSSAI-IDKYIGESARLIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSA 835
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
+++ + + +LL +LDGF++ V ++ T +D AL RPGR+DR + P + R
Sbjct: 836 DREIQRTLMELLNQLDGFDQLGKVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNEQSR 1015
Query: 857 ENILYSAAKETMDDQLVEYVDWKKVAE 883
IL A ++Y K+AE
Sbjct: 1016MEILKIHAAGIAKHGEIDYEAVVKLAE 1096
>TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AAA-ATPase
subunit RPT3 {Arabidopsis thaliana}, partial (94%)
Length = 1575
Score = 102 bits (255), Expect = 7e-22
Identities = 61/212 (28%), Positives = 110/212 (51%), Gaps = 4/212 (1%)
Frame = +2
Query: 669 MKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKT 727
+ + +KP + N+ D K+EI E V L + ++++G PRGVL+ G GTGKT
Sbjct: 542 LSQSEKPDVTYNDIGGCDIQKQEIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKT 721
Query: 728 SLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAG 787
LA A+A + + + ++G+ VR++F+ A++ AP I+F+++ D A
Sbjct: 722 MLAKAVANHTTAAFIRVVGSEF-VQKYLGEGPRMVRDVFRLAKENAPAIIFIDEVDAIAT 898
Query: 788 VRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFH 847
R +++ + + +LL ++DGF++ V ++ T +D AL RPGR+DR
Sbjct: 899 ARFDAQTGADREVQRILMELLNQMDGFDQTVNVKVIMATNRADTLDPALLRPGRLDRKIE 1078
Query: 848 LQRPTQAERENIL-YSAAKETMDDQ--LVEYV 876
P + ++ + AK + D+ L +YV
Sbjct: 1079FPLPDRRQKRLVFQVCTAKMNLSDEVDLEDYV 1174
>BF647954 homologue to GP|11094192|db 26S proteasome regulatory particle
triple-A ATPase subunit4 {Oryza sativa (japonica
cultivar-group)}, partial (53%)
Length = 649
Score = 99.4 bits (246), Expect = 7e-21
Identities = 59/183 (32%), Positives = 98/183 (53%)
Frame = +2
Query: 701 NPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSAS 760
NP F +G + P+GVL+ G GTGKT LA AIA+ +++ + + ++G+S+
Sbjct: 5 NPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASNIDANFLKVVSSAI-IDKYIGESSR 181
Query: 761 NVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGV 820
+RE+F ARD P I+F+++ D G R + +++ + + +LL +LDGF++ V
Sbjct: 182 LIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLMELLNQLDGFDQLGKV 361
Query: 821 VLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKK 880
++ T +D AL RPGR+DR + P + R IL A ++Y K
Sbjct: 362 KIIMATNRPDVLDPALLRPGRLDRKIEIPLPNEQSRMEILKIHAAGIAKHGEIDYEAVVK 541
Query: 881 VAE 883
+AE
Sbjct: 542 LAE 550
>TC90687 similar to GP|9759220|dbj|BAB09632.1 cell division protein FtsH
{Arabidopsis thaliana}, partial (28%)
Length = 813
Score = 99.0 bits (245), Expect = 1e-20
Identities = 51/114 (44%), Positives = 71/114 (61%)
Frame = +2
Query: 673 KKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMA 732
K I + + +D KEE+ E+V FL+NP + +GAR PRGVL+VG GTGKT LA A
Sbjct: 380 KGETITFADVAGVDEAKEELEEIVEFLRNPDRYARLGARPPRGVLLVGLPGTGKTLLAKA 559
Query: 733 IAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFA 786
+A EA VP + A + ++VG AS VR+LF A+ AP I+F+++ D A
Sbjct: 560 VAGEADVPFISCSASEF-VELYVGMGASRVRDLFARAKKEAPSIIFIDEIDAVA 718
>TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (39%)
Length = 1514
Score = 93.2 bits (230), Expect = 5e-19
Identities = 76/259 (29%), Positives = 123/259 (47%), Gaps = 17/259 (6%)
Frame = +1
Query: 633 NRRKLEQMAYYFDERKGRMRDRRREG-------VDPIKTAFEHMKRVKKPP-----IPLN 680
+ +K ++ + D RK + ++ +++G D AFE R + P + +
Sbjct: 661 DNKKTKKESKRDDSRKEKPKESKKDGDIKASAKSDSPDNAFEECIRQELIPANEIKVTFS 840
Query: 681 NFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAP-RGVLIVGERGTGKTSLAMAIAAEAK 738
+ ++D +KE + E V L+ P F+ G P +GVL+ G GTGKT LA AIA EA
Sbjct: 841 DIGALDDVKESLQEAVMLPLRRPDLFKGDGVLKPCKGVLLFGPPGTGKTMLAKAIANEAG 1020
Query: 739 VPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQ 798
+ + + + MW GQS NVR LF A +AP I+F+++ D G R E+
Sbjct: 1021ASFINVSPSTITS-MWQGQSEKNVRALFSLAAKVAPTIIFIDEVDSMLGQRSS--TREHS 1191
Query: 799 DHEAFINQLLVELDGF--EKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N+ + DG + + + ++A T +DEA+ R R R + P+ R
Sbjct: 1192SMRRVKNEFMSRWDGLLSKPDEKITVLAATNMPFDLDEAIIR--RFQRRIMVGLPSADNR 1365
Query: 857 ENILYS-AAKETMDDQLVE 874
E IL + AKE ++ E
Sbjct: 1366ETILKTILAKEKSENMNFE 1422
>TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana},
partial (77%)
Length = 1564
Score = 90.9 bits (224), Expect = 3e-18
Identities = 79/290 (27%), Positives = 137/290 (47%), Gaps = 11/290 (3%)
Frame = +1
Query: 594 IMLFIARAAISGFVLINVFQFMRRKIPRLLGYGPIQKNPNR---RKLEQMAYYFDERKGR 650
++L+ ARAA+S +VL+ +++ +PNR +K + +R GR
Sbjct: 193 LLLYAARAALSYWVLVTGLRYL---------------DPNRESSKKALEQKKEIAKRLGR 327
Query: 651 MRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMG 709
+ D I + + + ++ ++++K+ + E+V LQ P F
Sbjct: 328 PLIQTNSYEDVIACDVINPDHID---VEFDSIGGLETIKQTLFELVILPLQRPDLFSHGK 498
Query: 710 ARAP-RGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQT 768
P +GVL+ G GTGKT LA AIA E+ + ++ L W G + V +F
Sbjct: 499 LLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFINVRISNL-MSKWFGDAQKLVAAVFSL 675
Query: 769 ARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFIN---QLLVELDGF--EKQDGVVLM 823
A L P I+F+++ D F G R + DHEA +N + + DGF ++ V+++
Sbjct: 676 AHKLQPSIIFIDEVDSFLGQR------RSSDHEAVLNMKTEFMALWDGFATDQSARVMVL 837
Query: 824 ATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAK-ETMDDQL 872
A T ++DEA+ R R+ + F + P + ER +IL K E ++D +
Sbjct: 838 AATNRPSELDEAILR--RLPQAFEIGYPDRKERADILKVILKGEKVEDNI 981
>TC81426 similar to GP|6016719|gb|AAF01545.1| putative cell division control
protein {Arabidopsis thaliana}, partial (21%)
Length = 965
Score = 89.7 bits (221), Expect = 6e-18
Identities = 57/157 (36%), Positives = 83/157 (52%)
Frame = +2
Query: 718 IVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVIL 777
+ G G GKT +A A+A EA + IK +L +VG+S VR LF AR AP +L
Sbjct: 2 LFGPPGCGKTLIAKAVANEAGANFIHIKGPEL-LNKYVGESELAVRTLFNRARTCAPCVL 178
Query: 778 FVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQ 837
F ++ D RGK E +NQLL+ELDG E++ GV ++ T +D AL
Sbjct: 179 FFDEVDALTTKRGK---EGGWVIERLLNQLLIELDGAEQRRGVFVIGATNRPDVMDPALL 349
Query: 838 RPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVE 874
RPGR ++ ++ P+ +R IL + A+ D V+
Sbjct: 350 RPGRFGKLLYVPLPSPDDRVLILKALARNKHIDSSVD 460
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.332 0.143 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,909,964
Number of Sequences: 36976
Number of extensions: 519382
Number of successful extensions: 4220
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 3138
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 922
Number of HSP's gapped (non-prelim): 3441
length of query: 1184
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1077
effective length of database: 5,058,295
effective search space: 5447783715
effective search space used: 5447783715
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135461.15


