

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.13 + phase: 0
(560 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
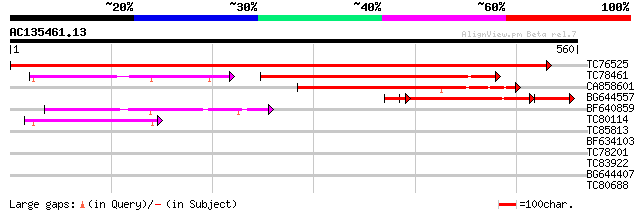
Score E
Sequences producing significant alignments: (bits) Value
TC76525 similar to GP|15450864|gb|AAK96703.1 Unknown protein {Ar... 1131 0.0
TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown prot... 280 e-118
CA858601 weakly similar to GP|22136978|gb unknown protein {Arabi... 240 9e-64
BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativ... 166 6e-53
BF640859 weakly similar to GP|15450387|gb At2g44230/F4I1.4 {Arab... 156 2e-38
TC80114 weakly similar to GP|6721151|gb|AAF26779.1| unknown prot... 135 5e-32
TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cy... 31 1.1
BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc... 31 1.4
TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-li... 30 2.5
TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180 ... 30 3.2
BG644407 similar to GP|12697612|dbj contains ESTs C97999(C0353) ... 29 4.2
TC80688 similar to GP|10177579|dbj|BAB10810. gene_id:K11I1.1~sim... 28 9.3
>TC76525 similar to GP|15450864|gb|AAK96703.1 Unknown protein {Arabidopsis
thaliana}, partial (75%)
Length = 2266
Score = 1131 bits (2925), Expect = 0.0
Identities = 534/535 (99%), Positives = 534/535 (99%)
Frame = +1
Query: 1 MFGFECFCWDSFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVW 60
MFGFECFCWDSFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVW
Sbjct: 661 MFGFECFCWDSFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVW 840
Query: 61 RCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESE 120
RCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESE
Sbjct: 841 RCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESE 1020
Query: 121 SPALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDL 180
SPALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDL
Sbjct: 1021 SPALKKPLNYSLIWCMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDL 1200
Query: 181 TEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLK 240
TEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLK
Sbjct: 1201 TEVCETSDLLLTIKSKKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLK 1380
Query: 241 NLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAI 300
NLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAI
Sbjct: 1381 NLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAI 1560
Query: 301 DYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWV 360
DYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWV
Sbjct: 1561 DYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWV 1740
Query: 361 FCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAF 420
FCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAF
Sbjct: 1741 FCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAF 1920
Query: 421 DLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVA 480
DLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVA
Sbjct: 1921 DLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVA 2100
Query: 481 AEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELF 535
AEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPI VRFSVENLFELF
Sbjct: 2101 AEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPILVRFSVENLFELF 2265
>TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown protein
{Arabidopsis thaliana}, partial (65%)
Length = 1464
Score = 280 bits (716), Expect(2) = e-118
Identities = 130/239 (54%), Positives = 172/239 (71%), Gaps = 2/239 (0%)
Frame = +3
Query: 248 AMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTN 306
+MPNL QI AL + + P +Y HP+EK+ P S+ W+F NGA+LY G ID G+N
Sbjct: 753 SMPNLPQIKALAQAHSPFMYLHPNEKFQPCSIKWYFTNGALLYKKGEESNPMEIDPLGSN 932
Query: 307 LPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNG 366
LP GG NDGA+W+DLP D+D R +KKG+ ++ + Y HVKP GG FTD+AMW+F PFNG
Sbjct: 933 LPQGGSNDGAYWLDLPKDKDNRERVKKGDFKNCQGYFHVKPIFGGTFTDLAMWIFYPFNG 1112
Query: 367 PATLKVSLM-NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFI 425
P TLKV L+ NI + KIGEH+GDWEH TLRVSNF GEL V+FS+HS G+WV++ ++EF
Sbjct: 1113PGTLKVGLIDNISLGKIGEHIGDWEHVTLRVSNFNGELKRVYFSQHSNGQWVDSSEVEFQ 1292
Query: 426 KENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYL 484
NKP+ YSS +GHA YP AG LQG+S IG++N+ KS+ ++D ++IV+ EYL
Sbjct: 1293SGNKPVAYSSLNGHAIYPKAGLVLQGTS--DIGIKNETKKSDLVIDFGVNFEIVSGEYL 1463
Score = 162 bits (410), Expect(2) = e-118
Identities = 85/213 (39%), Positives = 122/213 (56%), Gaps = 10/213 (4%)
Frame = +1
Query: 20 PLP----FSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFY 75
PLP F LP+ +P WPQGGGF G I LG+++V++++ F K+W LG TF+
Sbjct: 67 PLPIDTTFKLPANIPSWPQGGGFGNGIIDLGELKVSQISTFNKIWTTLEGGQGDLGATFF 246
Query: 76 RPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWC 135
P IP GF LG+Y N++PL G VLVA+ + + ALKKP++Y+L+W
Sbjct: 247 EPTGIPQGFFTLGHYSQPNNKPLFGWVLVAK-----------DESNGALKKPIDYTLVWS 393
Query: 136 MDS----HDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLL 191
S D+ Y WLP P GY +G +VTT P++P + ++CVR+DLT+ CE + +
Sbjct: 394 SKSEKIKQDKDGYIWLPIAPNGYSPLGHIVTTTPEKPSLDRIQCVRSDLTDQCEINSWIW 573
Query: 192 TIKSK--KNSFQVWNTQPCDRGMLARGVSVGTF 222
K + F V +P +RG+ A V VGTF
Sbjct: 574 GKDKKIDEKGFNVHYVRPINRGIEAPSVLVGTF 672
>CA858601 weakly similar to GP|22136978|gb unknown protein {Arabidopsis
thaliana}, partial (33%)
Length = 695
Score = 240 bits (613), Expect = 9e-64
Identities = 120/225 (53%), Positives = 149/225 (65%), Gaps = 5/225 (2%)
Frame = +2
Query: 285 NGAILYTAGNAKGKA-IDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYV 343
NGA+LY G I +GTNLP DGA+WIDLP D + +KKGN++SAE YV
Sbjct: 5 NGALLYKKGEESNPVPIAQNGTNLPQDPNTDGAYWIDLPADAANKERVKKGNLQSAESYV 184
Query: 344 HVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGEL 403
HVKP GG FTDIAMWVF PFNGP KV +N+++ KIGEHVGDWEH TLRVSN G+L
Sbjct: 185 HVKPMYGGTFTDIAMWVFYPFNGPGRAKVEFLNVKLGKIGEHVGDWEHVTLRVSNLDGQL 364
Query: 404 WSVFFSEHSGGKWVNAFDLEFI----KENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGV 459
W V+FS+H+ G W+++ LEF +P+VY+S HGHASYPHAG L G K GIG
Sbjct: 365 WHVYFSQHNLGSWIDSSQLEFQDGIGATKRPVVYASLHGHASYPHAGLVLLG--KNGIGA 538
Query: 460 RNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPT 504
R+D K ++D ++ +V+AEYLG V EP WL Y + G T
Sbjct: 539 RDDTNKGKNVMDMG-KFVLVSAEYLGSEV-KEPAWLNYFKGMGST 667
>BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativa (japonica
cultivar-group)}, partial (22%)
Length = 693
Score = 166 bits (420), Expect(2) = 6e-53
Identities = 81/134 (60%), Positives = 104/134 (77%), Gaps = 1/134 (0%)
Frame = -2
Query: 386 VGDWEHFTLRVSNFTGE-LWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPH 444
+G HF NFT L + + HSGGKWV+A++LE+I NK IVY+S++GHASYPH
Sbjct: 638 IGSISHF--E*GNFTWRTLEYILLTGHSGGKWVDAYELEYIDGNKAIVYASKNGHASYPH 465
Query: 445 AGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPT 504
GTY+QGSSKLGIG+RNDA +SN +DSS Y+IVAA YLGD V+ EP WLQYMR+WGP
Sbjct: 464 PGTYIQGSSKLGIGIRNDARRSNLRVDSSVHYEIVAAAYLGD-VVKEPQWLQYMRQWGPK 288
Query: 505 IVYDSRSEIEKIID 518
IVYDS++E++KI++
Sbjct: 287 IVYDSKTELDKILN 246
Score = 60.1 bits (144), Expect(2) = 6e-53
Identities = 26/40 (65%), Positives = 31/40 (77%)
Frame = -3
Query: 519 MLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
+LP+ +R S NLF P EL GEEGPTGPKEK+NW+GDE
Sbjct: 244 LLPLRLRSSFGNLFRKLPVELYGEEGPTGPKEKNNWIGDE 125
Score = 41.6 bits (96), Expect = 8e-04
Identities = 14/26 (53%), Positives = 21/26 (79%)
Frame = -1
Query: 371 KVSLMNIEMNKIGEHVGDWEHFTLRV 396
K + ++ +K+GEH+GDWEHFTLR+
Sbjct: 690 KFGMKSMAFSKVGEHIGDWEHFTLRI 613
>BF640859 weakly similar to GP|15450387|gb At2g44230/F4I1.4 {Arabidopsis
thaliana}, partial (23%)
Length = 655
Score = 156 bits (394), Expect = 2e-38
Identities = 90/233 (38%), Positives = 129/233 (54%), Gaps = 7/233 (3%)
Frame = +2
Query: 35 GGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHSN 94
G GFA G I LG ++V++V+ F KVW + G T + P IP GFS LG Y N
Sbjct: 2 GSGFANGIIDLGGLQVSQVSTFNKVWATYDGGPDNQGATIFEPTGIPQGFSMLGCYSQPN 181
Query: 95 DQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWCMD----SHDECVYFWLPNP 150
++PL G+VLVA++ + S + + LKKP++Y+L+ + D Y WLP
Sbjct: 182 NKPLFGYVLVAKDVS-------SSTTNSTLKKPIDYTLVLNTSTIKVNQDTTCYIWLPIA 340
Query: 151 PKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSFQVWNTQPCDR 210
P GYKA+G VVT D+P +++ CVR+DLT+ CE+S + N F ++ +P +
Sbjct: 341 PDGYKALGHVVTATQDKPSLDKIMCVRSDLTDQCESSSWIW----GSNDFNFYDVRPINX 508
Query: 211 GMLARGVSVGTFFC---GTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIE 260
G A GV VG F GT + CLKNL+S+ MPNL QI A+++
Sbjct: 509 GTQAPGVHVGAFVAQNGGTNIPP----SISCLKNLNSISKIMPNLXQIDAILK 655
>TC80114 weakly similar to GP|6721151|gb|AAF26779.1| unknown protein
{Arabidopsis thaliana}, partial (12%)
Length = 765
Score = 135 bits (339), Expect = 5e-32
Identities = 69/156 (44%), Positives = 91/156 (58%), Gaps = 19/156 (12%)
Frame = +3
Query: 15 FFDSDPLP----------FSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWRCTN 64
FF S P P FSL +PLPQWPQG FA G ++LG+IEV K+ KFE VW
Sbjct: 297 FFPSKPNPSDHHHLLPNTFSLNTPLPQWPQGQDFASGIVNLGEIEVCKITKFETVWNSNV 476
Query: 65 SNGKALGFTFYRPLEIPDGFSCLGYYCHSNDQPLRGHVLVARETTSKSQAD-CSESESPA 123
TFY+P+ IPDGF LG+YC + +PL G VL A++ S + C++++ PA
Sbjct: 477 MVEPRKAITFYKPVGIPDGFHILGHYCQPSYKPLWGFVLAAKQVADSSYNNICNQNKLPA 656
Query: 124 LKKPLNYSLIWCMDSH--------DECVYFWLPNPP 151
L+ PL+++LIWC +S D YFWLP PP
Sbjct: 657 LRNPLDFALIWCTNSGRKKIAMPVDSAAYFWLPQPP 764
>TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cytochrome P450
reductase {Pisum sativum}, complete
Length = 2581
Score = 31.2 bits (69), Expect = 1.1
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 1/73 (1%)
Frame = +3
Query: 234 VDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTA 292
VD + L+ +H L+++ GPT + + +S W GA +Y
Sbjct: 1887 VDYIYEDELNHFVHGGA-LSELIVAFSREGPTKEYVQHKMIEKASDIWNMISQGAYIYVC 2063
Query: 293 GNAKGKAIDYHGT 305
G+AKG A D H T
Sbjct: 2064 GDAKGMAKDVHRT 2102
>BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc finger
protein {Arabidopsis thaliana}, partial (3%)
Length = 369
Score = 30.8 bits (68), Expect = 1.4
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Frame = -1
Query: 238 CLKNLDSLLHAMPNLNQIHALIE-----HYGPTVYFHPDEKYMPSSVSW 281
C+ LDS H + L Q+H L E H+ T + HP+++ M W
Sbjct: 198 CVDRLDSTDHVVEYLIQMHILYEMPLGIHHRETNFHHPNKQCMQDHYIW 52
>TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 2047
Score = 30.0 bits (66), Expect = 2.5
Identities = 19/69 (27%), Positives = 32/69 (45%)
Frame = -3
Query: 466 SNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVR 525
+NFI+D+S R+ +V L + + Y + W P + K + L V
Sbjct: 1685 TNFIVDNSSRWDVVRRIGLEESCVVVLLSNNYHKFWSPIFI--------KKLACLNDLVE 1530
Query: 526 FSVENLFEL 534
F+++NL EL
Sbjct: 1529 FNIQNLIEL 1503
>TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180
{Arabidopsis thaliana}, partial (21%)
Length = 619
Score = 29.6 bits (65), Expect = 3.2
Identities = 18/49 (36%), Positives = 22/49 (44%)
Frame = -3
Query: 11 SFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKV 59
SFP PLP P PLP P F IE++K+NK K+
Sbjct: 194 SFPLPLPLLPLPLPFPLPLPLLPLPLPFPWP*AMAASIEISKINKATKM 48
>BG644407 similar to GP|12697612|dbj contains ESTs C97999(C0353)
D15249(C0353)~similar to Arabidopsis thaliana chromosome
2 At2g01100~, partial (21%)
Length = 764
Score = 29.3 bits (64), Expect = 4.2
Identities = 13/28 (46%), Positives = 16/28 (56%)
Frame = -3
Query: 4 FECFCWDSFPEFFDSDPLPFSLPSPLPQ 31
F CF WD +FF S+ L SLP P+
Sbjct: 465 FLCFLWDFLVDFFFSNSLSVSLPLSDPE 382
>TC80688 similar to GP|10177579|dbj|BAB10810. gene_id:K11I1.1~similar to
unknown protein~sp|P74035 {Arabidopsis thaliana},
partial (13%)
Length = 664
Score = 28.1 bits (61), Expect = 9.3
Identities = 11/20 (55%), Positives = 13/20 (65%)
Frame = +3
Query: 11 SFPEFFDSDPLPFSLPSPLP 30
SF FF + LP+S P PLP
Sbjct: 132 SFQTFFSNQTLPYSFPFPLP 191
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,044,651
Number of Sequences: 36976
Number of extensions: 329169
Number of successful extensions: 1648
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1614
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1635
length of query: 560
length of database: 9,014,727
effective HSP length: 101
effective length of query: 459
effective length of database: 5,280,151
effective search space: 2423589309
effective search space used: 2423589309
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135461.13


