

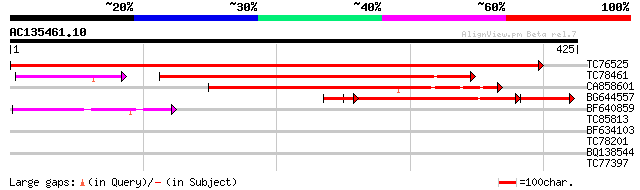
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.10 + phase: 0
(425 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76525 similar to GP|15450864|gb|AAK96703.1 Unknown protein {Ar... 838 0.0
TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown prot... 280 4e-90
CA858601 weakly similar to GP|22136978|gb unknown protein {Arabi... 240 7e-64
BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativ... 166 4e-53
BF640859 weakly similar to GP|15450387|gb At2g44230/F4I1.4 {Arab... 91 1e-18
TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cy... 31 0.79
BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc... 31 1.0
TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-li... 30 1.8
BQ138544 similar to GP|8777399|dbj| emb|CAB86483.1~gene_id:MFB16... 28 8.8
TC77397 homologue to PIR|S37159|S37159 NADPH--ferrihemoprotein r... 28 8.8
>TC76525 similar to GP|15450864|gb|AAK96703.1 Unknown protein {Arabidopsis
thaliana}, partial (75%)
Length = 2266
Score = 838 bits (2165), Expect = 0.0
Identities = 399/400 (99%), Positives = 399/400 (99%)
Frame = +1
Query: 1 MDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKS 60
MDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKS
Sbjct: 1066 MDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKS 1245
Query: 61 KKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQI 120
KKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQI
Sbjct: 1246 KKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQI 1425
Query: 121 HALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDG 180
HALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDG
Sbjct: 1426 HALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDG 1605
Query: 181 AFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLM 240
AFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLM
Sbjct: 1606 AFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLM 1785
Query: 241 NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSS 300
NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSS
Sbjct: 1786 NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSS 1965
Query: 301 RHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWL 360
RHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWL
Sbjct: 1966 RHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWL 2145
Query: 361 QYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELF 400
QYMREWGPTIVYDSRSEIEKIIDMLPI VRFSVENLFELF
Sbjct: 2146 QYMREWGPTIVYDSRSEIEKIIDMLPILVRFSVENLFELF 2265
>TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown protein
{Arabidopsis thaliana}, partial (65%)
Length = 1464
Score = 280 bits (716), Expect(2) = 4e-90
Identities = 130/239 (54%), Positives = 172/239 (71%), Gaps = 2/239 (0%)
Frame = +3
Query: 113 AMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTN 171
+MPNL QI AL + + P +Y HP+EK+ P S+ W+F NGA+LY G ID G+N
Sbjct: 753 SMPNLPQIKALAQAHSPFMYLHPNEKFQPCSIKWYFTNGALLYKKGEESNPMEIDPLGSN 932
Query: 172 LPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNG 231
LP GG NDGA+W+DLP D+D R +KKG+ ++ + Y HVKP GG FTD+AMW+F PFNG
Sbjct: 933 LPQGGSNDGAYWLDLPKDKDNRERVKKGDFKNCQGYFHVKPIFGGTFTDLAMWIFYPFNG 1112
Query: 232 PATLKVSLM-NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFI 290
P TLKV L+ NI + KIGEH+GDWEH TLRVSNF GEL V+FS+HS G+WV++ ++EF
Sbjct: 1113PGTLKVGLIDNISLGKIGEHIGDWEHVTLRVSNFNGELKRVYFSQHSNGQWVDSSEVEFQ 1292
Query: 291 KENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYL 349
NKP+ YSS +GHA YP AG LQG+S IG++N+ KS+ ++D ++IV+ EYL
Sbjct: 1293SGNKPVAYSSLNGHAIYPKAGLVLQGTS--DIGIKNETKKSDLVIDFGVNFEIVSGEYL 1463
Score = 69.7 bits (169), Expect(2) = 4e-90
Identities = 34/85 (40%), Positives = 50/85 (58%), Gaps = 2/85 (2%)
Frame = +1
Query: 5 DECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSK--K 62
D+ Y WLP P GY +G +VTT P++P + ++CVR+DLT+ CE + + K +
Sbjct: 418 DKDGYIWLPIAPNGYSPLGHIVTTTPEKPSLDRIQCVRSDLTDQCEINSWIWGKDKKIDE 597
Query: 63 NSFQVWNTQPCDRGMLARGVSVGTF 87
F V +P +RG+ A V VGTF
Sbjct: 598 KGFNVHYVRPINRGIEAPSVLVGTF 672
>CA858601 weakly similar to GP|22136978|gb unknown protein {Arabidopsis
thaliana}, partial (33%)
Length = 695
Score = 240 bits (613), Expect = 7e-64
Identities = 120/225 (53%), Positives = 149/225 (65%), Gaps = 5/225 (2%)
Frame = +2
Query: 150 NGAILYTAGNAKGKA-IDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYV 208
NGA+LY G I +GTNLP DGA+WIDLP D + +KKGN++SAE YV
Sbjct: 5 NGALLYKKGEESNPVPIAQNGTNLPQDPNTDGAYWIDLPADAANKERVKKGNLQSAESYV 184
Query: 209 HVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGEL 268
HVKP GG FTDIAMWVF PFNGP KV +N+++ KIGEHVGDWEH TLRVSN G+L
Sbjct: 185 HVKPMYGGTFTDIAMWVFYPFNGPGRAKVEFLNVKLGKIGEHVGDWEHVTLRVSNLDGQL 364
Query: 269 WSVFFSEHSGGKWVNAFDLEFI----KENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGV 324
W V+FS+H+ G W+++ LEF +P+VY+S HGHASYPHAG L G K GIG
Sbjct: 365 WHVYFSQHNLGSWIDSSQLEFQDGIGATKRPVVYASLHGHASYPHAGLVLLG--KNGIGA 538
Query: 325 RNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPT 369
R+D K ++D ++ +V+AEYLG V EP WL Y + G T
Sbjct: 539 RDDTNKGKNVMDMG-KFVLVSAEYLGSEV-KEPAWLNYFKGMGST 667
>BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativa (japonica
cultivar-group)}, partial (22%)
Length = 693
Score = 166 bits (420), Expect(2) = 4e-53
Identities = 81/134 (60%), Positives = 104/134 (77%), Gaps = 1/134 (0%)
Frame = -2
Query: 251 VGDWEHFTLRVSNFTGE-LWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPH 309
+G HF NFT L + + HSGGKWV+A++LE+I NK IVY+S++GHASYPH
Sbjct: 638 IGSISHF--E*GNFTWRTLEYILLTGHSGGKWVDAYELEYIDGNKAIVYASKNGHASYPH 465
Query: 310 AGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPT 369
GTY+QGSSKLGIG+RNDA +SN +DSS Y+IVAA YLGD V+ EP WLQYMR+WGP
Sbjct: 464 PGTYIQGSSKLGIGIRNDARRSNLRVDSSVHYEIVAAAYLGD-VVKEPQWLQYMRQWGPK 288
Query: 370 IVYDSRSEIEKIID 383
IVYDS++E++KI++
Sbjct: 287 IVYDSKTELDKILN 246
Score = 60.1 bits (144), Expect(2) = 4e-53
Identities = 26/40 (65%), Positives = 31/40 (77%)
Frame = -3
Query: 384 MLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 423
+LP+ +R S NLF P EL GEEGPTGPKEK+NW+GDE
Sbjct: 244 LLPLRLRSSFGNLFRKLPVELYGEEGPTGPKEKNNWIGDE 125
Score = 41.6 bits (96), Expect = 6e-04
Identities = 14/26 (53%), Positives = 21/26 (79%)
Frame = -1
Query: 236 KVSLMNIEMNKIGEHVGDWEHFTLRV 261
K + ++ +K+GEH+GDWEHFTLR+
Sbjct: 690 KFGMKSMAFSKVGEHIGDWEHFTLRI 613
>BF640859 weakly similar to GP|15450387|gb At2g44230/F4I1.4 {Arabidopsis
thaliana}, partial (23%)
Length = 655
Score = 90.5 bits (223), Expect = 1e-18
Identities = 50/126 (39%), Positives = 70/126 (54%), Gaps = 3/126 (2%)
Frame = +2
Query: 3 SHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKK 62
+ D Y WLP P GYKA+G VVT D+P +++ CVR+DLT+ CE+S +
Sbjct: 302 NQDTTCYIWLPIAPDGYKALGHVVTATQDKPSLDKIMCVRSDLTDQCESSSWIW----GS 469
Query: 63 NSFQVWNTQPCDRGMLARGVSVGTFFC---GTYFDSEQVVDVVCLKNLDSLLHAMPNLNQ 119
N F ++ +P + G A GV VG F GT + CLKNL+S+ MPNL Q
Sbjct: 470 NDFNFYDVRPINXGTQAPGVHVGAFVAQNGGTNIPP----SISCLKNLNSISKIMPNLXQ 637
Query: 120 IHALIE 125
I A+++
Sbjct: 638 IDAILK 655
>TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cytochrome P450
reductase {Pisum sativum}, complete
Length = 2581
Score = 31.2 bits (69), Expect = 0.79
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 1/73 (1%)
Frame = +3
Query: 99 VDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTA 157
VD + L+ +H L+++ GPT + + +S W GA +Y
Sbjct: 1887 VDYIYEDELNHFVHGGA-LSELIVAFSREGPTKEYVQHKMIEKASDIWNMISQGAYIYVC 2063
Query: 158 GNAKGKAIDYHGT 170
G+AKG A D H T
Sbjct: 2064 GDAKGMAKDVHRT 2102
>BF634103 weakly similar to GP|20260434|gb putative PHD-type zinc finger
protein {Arabidopsis thaliana}, partial (3%)
Length = 369
Score = 30.8 bits (68), Expect = 1.0
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Frame = -1
Query: 103 CLKNLDSLLHAMPNLNQIHALIE-----HYGPTVYFHPDEKYMPSSVSW 146
C+ LDS H + L Q+H L E H+ T + HP+++ M W
Sbjct: 198 CVDRLDSTDHVVEYLIQMHILYEMPLGIHHRETNFHHPNKQCMQDHYIW 52
>TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 2047
Score = 30.0 bits (66), Expect = 1.8
Identities = 19/69 (27%), Positives = 32/69 (45%)
Frame = -3
Query: 331 SNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVR 390
+NFI+D+S R+ +V L + + Y + W P + K + L V
Sbjct: 1685 TNFIVDNSSRWDVVRRIGLEESCVVVLLSNNYHKFWSPIFI--------KKLACLNDLVE 1530
Query: 391 FSVENLFEL 399
F+++NL EL
Sbjct: 1529 FNIQNLIEL 1503
>BQ138544 similar to GP|8777399|dbj| emb|CAB86483.1~gene_id:MFB16.16~similar
to unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 395
Score = 27.7 bits (60), Expect = 8.8
Identities = 12/42 (28%), Positives = 23/42 (54%)
Frame = -1
Query: 92 YFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYF 133
Y S+ + ++C +L L+ AMP+++ + Y P +YF
Sbjct: 182 YSYSKVMTSIICYCHLHVLMIAMPSISYSSYIFPLYCPLIYF 57
>TC77397 homologue to PIR|S37159|S37159 NADPH--ferrihemoprotein reductase (EC
1.6.2.4) - spring vetch, complete
Length = 2519
Score = 27.7 bits (60), Expect = 8.8
Identities = 16/56 (28%), Positives = 24/56 (42%), Gaps = 1/56 (1%)
Frame = +2
Query: 116 NLNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTAGNAKGKAIDYHGT 170
+L+++ GP + + +S W G LY G+AKG A D H T
Sbjct: 1889 SLSELIVAFSREGPEKEYVQHKMMDKASYFWSLISQGGYLYVCGDAKGMARDVHRT 2056
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,156,024
Number of Sequences: 36976
Number of extensions: 221358
Number of successful extensions: 938
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 924
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 930
length of query: 425
length of database: 9,014,727
effective HSP length: 99
effective length of query: 326
effective length of database: 5,354,103
effective search space: 1745437578
effective search space used: 1745437578
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135461.10


