

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135415.3 + phase: 0 /pseudo
(436 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
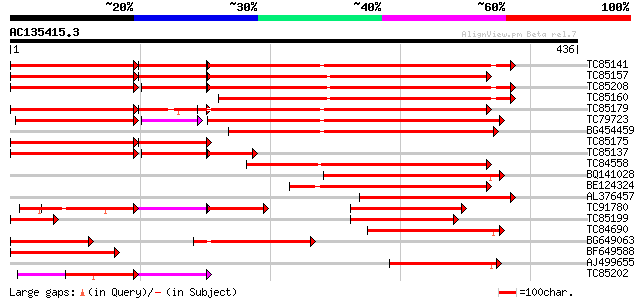
Score E
Sequences producing significant alignments: (bits) Value
TC85141 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 313 8e-86
TC85157 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 309 2e-84
TC85208 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 305 2e-83
TC85160 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 295 2e-80
TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza s... 280 8e-76
TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-al... 266 9e-72
BG454459 similar to SP|P34823|EF12 Elongation factor 1-alpha (EF... 253 1e-67
TC85175 homologue to GP|3869088|dbj|BAA34348.1 elongation factor... 156 6e-56
TC85137 GP|3869088|dbj|BAA34348.1 elongation factor-1 alpha {Nic... 156 2e-51
TC84558 weakly similar to SP|Q04634|EF1A_TETPY Elongation factor... 191 4e-49
BQ141028 homologue to SP|P40911|EF1A Elongation factor 1-alpha (... 154 6e-38
BE124324 weakly similar to SP|Q41011|EF1A Elongation factor 1-al... 150 1e-36
AL376457 similar to SP|P13549|EF10 Elongation factor 1-alpha so... 139 2e-33
TC91780 homologue to SP|P17786|EF1A_LYCES Elongation factor 1-al... 129 2e-30
TC85199 homologue to SP|O24534|EF1A_VICFA Elongation factor 1-al... 125 3e-29
TC84690 similar to GP|21540004|gb|AAM54368.1 elongation factor 1... 125 4e-29
BG649063 homologue to PIR|F86214|F86 protein T6D22.2 [imported] ... 125 4e-29
BF649588 similar to SP|P29521|EF11_ Elongation factor 1-alpha (E... 120 8e-28
AJ499655 homologue to SP|P28295|EF1A_ Elongation factor 1-alpha ... 104 8e-23
TC85202 homologue to GP|18873727|gb|AAL79775.1 elongation factor... 100 8e-22
>TC85141 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 1881
Score = 313 bits (802), Expect = 8e-86
Identities = 159/237 (67%), Positives = 188/237 (79%)
Frame = +3
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG +KPGMV+TFAPT LQ+
Sbjct: 723 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGTMKPGMVVTFAPTGLQT 902
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N +S K+L RGYVASN++D+PA EAA FTS+VI
Sbjct: 903 EVKSVEMHHESLTEALPGDNVGFNVKN--VSVKDLKRGYVASNTKDDPAKEAANFTSQVI 1076
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAGI+KMI
Sbjct: 1077IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKEIEKEPKFLKNGDAGIIKMI 1256
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI V+K K K+T S +K
Sbjct: 1257PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKGVEK---KEPGTAKITKSAAKKK 1418
Score = 156 bits (395), Expect(2) = 8e-56
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +3
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 78 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 257
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 258 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 374
Score = 79.0 bits (193), Expect(2) = 8e-56
Identities = 37/56 (66%), Positives = 41/56 (73%)
Frame = +2
Query: 100 N*EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
+*EHD+ + S*LCCS H FH+ F SW GWSD TCS C HSWCEANDLLLQQ
Sbjct: 371 H*EHDYWYIPS*LCCSHH*FHHWWF*SWYFQGWSDS*TCSPCFHSWCEANDLLLQQ 538
>TC85157 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 1738
Score = 309 bits (791), Expect = 2e-84
Identities = 154/218 (70%), Positives = 179/218 (81%)
Frame = +1
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+TFAPT L +
Sbjct: 727 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVITFAPTGLTT 906
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RGYVASNS+D+PA EAA FTS+VI
Sbjct: 907 EVKSVEMHHEALTEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKEAANFTSQVI 1080
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAGI+KMI
Sbjct: 1081IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKEIEKEPKFLKNGDAGIIKMI 1260
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK 370
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI +V+K
Sbjct: 1261PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKAVEK 1374
Score = 156 bits (395), Expect(2) = 6e-56
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 82 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 261
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 262 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 378
Score = 79.3 bits (194), Expect(2) = 6e-56
Identities = 39/56 (69%), Positives = 42/56 (74%)
Frame = +3
Query: 100 N*EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
+*EHDHR++ S*LC S H FHY F S *GWSD TCS C HSWCEANDLLLQQ
Sbjct: 375 H*EHDHRNIPS*LCRSHH*FHYWWF*SRYF*GWSDP*TCSPCFHSWCEANDLLLQQ 542
>TC85208 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 2287
Score = 305 bits (781), Expect = 2e-83
Identities = 154/237 (64%), Positives = 185/237 (77%)
Frame = +2
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TF PT L +
Sbjct: 1118 LLEALDQINEPKRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGIVKPGMVVTFGPTGLTT 1297
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 1298 EVKSVEMHHEALTEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVI 1471
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAGI+KM+
Sbjct: 1472 IMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKEIEKEPKFLKNGDAGIIKMV 1651
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE EYP LGRFAVRDMRQTVAVGVI +V+K K+ K+T + +K
Sbjct: 1652 PTKPMVVETFAEYPPLGRFAVRDMRQTVAVGVIKAVEK---KDPTGAKVTKAAAKKK 1813
Score = 156 bits (394), Expect(2) = 8e-49
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +2
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 473 MGKEKIHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 652
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 653 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 769
Score = 55.8 bits (133), Expect(2) = 8e-49
Identities = 27/54 (50%), Positives = 37/54 (68%)
Frame = +1
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
EHD+ +L *LC + H H+ F SW +*GW+D S+C++SW EA+DLLL Q
Sbjct: 772 EHDYWNLPG*LCRAYH*LHHWWF*SWYL*GWTDP*ARSSCIYSWSEADDLLL*Q 933
>TC85160 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (24%)
Length = 1198
Score = 295 bits (756), Expect = 2e-80
Identities = 149/229 (65%), Positives = 178/229 (77%)
Frame = +3
Query: 161 SEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQSGVKSIQKF 220
+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TF PT L + VKS++
Sbjct: 6 NEPKRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGIVKPGMVVTFGPTGLTTEVKSVEMH 185
Query: 221 QENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVIIINHPSVI 280
E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VII+NHP I
Sbjct: 186 HEALTEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVIIMNHPGQI 359
Query: 281 TKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVE 340
GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAGI+KM+P KPMVVE
Sbjct: 360 GNGYAPVLDCHTSHIAVKFAELVTKIDRRSGKEIEKEPKFLKNGDAGIIKMVPTKPMVVE 539
Query: 341 GINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
EYP LGRFAVRDMRQTVAVGVI +V+K K+ K+T + +K
Sbjct: 540 TFAEYPPLGRFAVRDMRQTVAVGVIKAVEK---KDPTGAKVTKAAAKKK 677
>TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza sativa},
partial (96%)
Length = 1789
Score = 280 bits (716), Expect = 8e-76
Identities = 147/226 (65%), Positives = 171/226 (75%)
Frame = +2
Query: 145 WCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLT 204
W E LL +A+DQI+EP RP KPLRLPL+ VYKIGGIGTVPVG VETGVLKPGM++T
Sbjct: 785 WYEGPTLL--EALDQINEPNRPSNKPLRLPLRDVYKIGGIGTVPVGCVETGVLKPGMMVT 958
Query: 205 FAPTKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEA 264
FAP LQ+ VKS+Q N+ EALP D VGF + N +S ++L RGYV SNS+ +PA EA
Sbjct: 959 FAPIGLQAEVKSVQMDHTNLTEALPGDNVGFNVEN--VSAEHLKRGYVVSNSKHDPAKEA 1132
Query: 265 AKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNG 324
F S VII NHP I GYTP+L CHTSH AVKF KL K DR +G +E +PK LKNG
Sbjct: 1133 DNFISCVIIRNHPGQIGNGYTPVLHCHTSHTAVKFDKLLAKIDRCSGKVIENEPKFLKNG 1312
Query: 325 DAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK 370
DAGI+KMIP KPMVVE + YPSLGRFAVRD+R+TVAVGVI +VKK
Sbjct: 1313 DAGIIKMIPTKPMVVETFSNYPSLGRFAVRDIRRTVAVGVIKAVKK 1450
Score = 161 bits (408), Expect(2) = 2e-51
Identities = 80/99 (80%), Positives = 85/99 (85%)
Frame = +2
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K INIVFIGHV+SGKSTTAGHL+YKLGGI+K IE KEAAE+ M SFKYAWVL
Sbjct: 152 MDKGKVGINIVFIGHVDSGKSTTAGHLIYKLGGIKKDVIERFEKEAAEINMCSFKYAWVL 331
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDIS+ KFETTKY CTLIDAPGHRDFI
Sbjct: 332 DKLKAERERGITIDISLSKFETTKYYCTLIDAPGHRDFI 448
Score = 58.9 bits (141), Expect(2) = 2e-51
Identities = 33/62 (53%), Positives = 39/62 (62%), Gaps = 6/62 (9%)
Frame = +1
Query: 100 N*EHDHRDLRS*LCCSSHQFHYSRFASWL------V*GWSDLGTCSTCVHSWCEANDLLL 153
+*EHDHR++ S*LCCS F+Y W+ *G SD TC C + WCEANDLLL
Sbjct: 445 H*EHDHRNIPS*LCCSRI*FYYY----WI*L*NCYF*GSSDP*TCCPCFNPWCEANDLLL 612
Query: 154 QQ 155
QQ
Sbjct: 613 QQ 618
>TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-alpha
(EF-1-alpha). {Podospora anserina}, complete
Length = 1537
Score = 266 bits (681), Expect = 9e-72
Identities = 131/228 (57%), Positives = 166/228 (72%)
Frame = +2
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +AID I PKRP KPLRLPLQ VYKIGGIGTVPVGR+ETGVLKPGMV+TFAP+ + +
Sbjct: 755 LLEAIDAIEPPKRPTDKPLRLPLQDVYKIGGIGTVPVGRIETGVLKPGMVVTFAPSNVTT 934
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + E P D VGF + N +S K++ RG VA +S+++P AA F ++VI
Sbjct: 935 EVKSVEMHHEQLTEGQPGDNVGFNVKN--VSVKDIRRGNVAGDSKNDPPAGAASFNAQVI 1108
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
++NHP + GY P+LDCHT+H+A KF+++ K DR TG +E PK +K+GDA IVKM+
Sbjct: 1109VLNHPGQVGAGYAPVLDCHTAHIACKFSEILEKIDRRTGKSVENNPKFIKSGDAAIVKMV 1288
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPM VE +YP LGRFAVRDMRQTVAVGVI SV K+ + TK
Sbjct: 1289PSKPMCVEAFTDYPPLGRFAVRDMRQTVAVGVIKSVDKSAATSGKVTK 1432
Score = 141 bits (356), Expect(2) = 6e-36
Identities = 69/95 (72%), Positives = 78/95 (81%)
Frame = +1
Query: 5 KPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLK 64
K +IN+V IGHV+SGKSTT GHL+Y+ GGI K IE KEAAE+ SFKYAWVLDKLK
Sbjct: 91 KTHINVVVIGHVDSGKSTTTGHLIYQCGGIDKRTIEKFEKEAAELGKGSFKYAWVLDKLK 270
Query: 65 AERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
AERERGITIDI++ KFET KY T+IDAPGHRDFI
Sbjct: 271 AERERGITIDIALWKFETPKYYVTVIDAPGHRDFI 375
Score = 27.3 bits (59), Expect(2) = 6e-36
Identities = 17/47 (36%), Positives = 21/47 (44%)
Frame = +3
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEA 148
EHD+ L L S H Y W + GWSD S +H C+A
Sbjct: 378 EHDYWYLPGRLRRSHHCRWYW*VRGWYLQGWSDS*ARSARLHPGCQA 518
>BG454459 similar to SP|P34823|EF12 Elongation factor 1-alpha (EF-1-alpha).
[Carrot] {Daucus carota}, partial (46%)
Length = 642
Score = 253 bits (645), Expect = 1e-67
Identities = 126/208 (60%), Positives = 154/208 (73%)
Frame = +1
Query: 169 KPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQSGVKSIQKFQENINEAL 228
KPLRLPLQ VYKIGGIGTVPVGRVETGV+KPGMV+ FAP+ L + VKS++ E++ EA
Sbjct: 7 KPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVVVFAPSGLTTEVKSVEMHHESLPEAN 186
Query: 229 PADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVIIINHPSVITKGYTPIL 288
P D VGF + N +S K L RG+VAS+S+++PA EA +FT++VII+NHP I GY P+L
Sbjct: 187 PGDNVGFNVKN--VSVKELKRGFVASDSKNDPAKEATEFTAQVIIMNHPGQIGNGYAPVL 360
Query: 289 DCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSL 348
DCHT+H+A KFA + TK DR +G ELE PK +KNGDA VKM+P KPM VE YP L
Sbjct: 361 DCHTAHIACKFADITTKIDRRSGKELEASPKFIKNGDAAYVKMVPXKPMCVEPFTTYPPL 540
Query: 349 GRFAVRDMRQTVAVGVIMSVKKNGYKNK 376
GRF VRDMR TV VG+I +V K K
Sbjct: 541 GRFXVRDMRXTVXVGIIKAVXKKDTTGK 624
>TC85175 homologue to GP|3869088|dbj|BAA34348.1 elongation factor-1 alpha
{Nicotiana paniculata}, partial (53%)
Length = 831
Score = 156 bits (395), Expect(2) = 6e-56
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +3
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 108 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 287
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 288 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 404
Score = 79.3 bits (194), Expect(2) = 6e-56
Identities = 39/56 (69%), Positives = 42/56 (74%)
Frame = +2
Query: 100 N*EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
+*EHDHR++ S*LC S H FHY F S *GWSD TCS C HSWCEANDLLLQQ
Sbjct: 401 H*EHDHRNIPS*LCRSHH*FHYWWF*SRYF*GWSDP*TCSPCFHSWCEANDLLLQQ 568
Score = 35.8 bits (81), Expect = 0.033
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = +3
Query: 156 AIDQISEPKRPEGKPLRLPL 175
A+DQI+EPK P KPLRLPL
Sbjct: 762 ALDQINEPKEPSDKPLRLPL 821
>TC85137 GP|3869088|dbj|BAA34348.1 elongation factor-1 alpha {Nicotiana
paniculata}, partial (56%)
Length = 1023
Score = 156 bits (394), Expect(2) = 2e-51
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 238 MGKEKIHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 417
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 418 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 534
Score = 66.6 bits (161), Expect = 2e-11
Identities = 31/38 (81%), Positives = 34/38 (88%)
Frame = +1
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVG 190
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVG
Sbjct: 883 LLEALDQINEPKRPTDKPLRLPLQDVYKIGGIGTVPVG 996
Score = 64.3 bits (155), Expect(2) = 2e-51
Identities = 29/54 (53%), Positives = 38/54 (69%)
Frame = +3
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
EHD+ +L *LCC H H+ F SW +*GW+D CS+C++SW EA+DLLL Q
Sbjct: 537 EHDYWNLPG*LCCPYH*LHHWWF*SWYL*GWTDP*ACSSCIYSWSEADDLLL*Q 698
>TC84558 weakly similar to SP|Q04634|EF1A_TETPY Elongation factor 1-alpha
(EF-1-alpha) (14-nm filament-associated protein).
{Tetrahymena pyriformis}, partial (42%)
Length = 611
Score = 191 bits (486), Expect = 4e-49
Identities = 92/188 (48%), Positives = 127/188 (66%)
Frame = +2
Query: 183 GIGTVPVGRVETGVLKPGMVLTFAPTKLQSGVKSIQKFQENINEALPADIVGFRLTNKTL 242
G+GTVPVGRVETG++K GM + F P K + K ++ E + EA P D VGF + K +
Sbjct: 5 GVGTVPVGRVETGIIKAGMQIAFTPGKQVAECKQVEMHHEVLPEAGPGDNVGFNV--KGI 178
Query: 243 SDKNLMRGYVASNSEDEPAMEAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKL 302
K L RG VAS++++ PA E F ++V+++NHP I GYTP++DCHT+H+A KF ++
Sbjct: 179 DSKELKRGNVASDAKNNPASEVTDFLAQVVVLNHPGNIAAGYTPVVDCHTAHIACKFQEI 358
Query: 303 ATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAV 362
TK D+ TG +E+ P +K GDA ++ + P KPMVVE + LGRFAVRDM+QTVAV
Sbjct: 359 KTKLDKRTGKVVEENPAHIKTGDAAMIVLTPTKPMVVEVYQNFAPLGRFAVRDMKQTVAV 538
Query: 363 GVIMSVKK 370
GVI + K
Sbjct: 539 GVIKEITK 562
>BQ141028 homologue to SP|P40911|EF1A Elongation factor 1-alpha (EF-1-alpha).
[Histoplasma capsulatum] {Ajellomyces capsulata},
partial (31%)
Length = 616
Score = 154 bits (389), Expect = 6e-38
Identities = 77/142 (54%), Positives = 96/142 (67%), Gaps = 3/142 (2%)
Frame = -2
Query: 242 LSDKNLMRGYVASNSEDEPAMEAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAK 301
+S K + RG VA +S+ +P F ++VI++NHP + GY P+LDCHT+H+A KF++
Sbjct: 612 VSVKEIRRGNVAGDSKQDPPKGXESFNAQVIVLNHPGQVGAGYAPVLDCHTAHIACKFSE 433
Query: 302 LATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVA 361
L K DR TG E PK +K+GDA IVKM+P KPM VE +YP LGRFAVRDMRQTVA
Sbjct: 432 LLEKIDRRTGKSTEASPKFIKSGDAAIVKMVPSKPMCVEAFTDYPPLGRFAVRDMRQTVA 253
Query: 362 VGVIMSV---KKNGYKNKAATK 380
VGVI SV K G KAA K
Sbjct: 252 VGVIKSVVKADKAGKVTKAAQK 187
>BE124324 weakly similar to SP|Q41011|EF1A Elongation factor 1-alpha
(EF-1-alpha). [Garden pea] {Pisum sativum}, partial
(32%)
Length = 625
Score = 150 bits (378), Expect = 1e-36
Identities = 81/155 (52%), Positives = 104/155 (66%)
Frame = +3
Query: 216 SIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVIIIN 275
S + +E ++ A P DIV F N +S +L +GYVASN ED+PA E FTS VII N
Sbjct: 12 SSEMHREPLDVAYPGDIVRF---NVNVSATDLKQGYVASNPEDDPAKEVTHFTSHVIITN 182
Query: 276 HPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMK 335
HPS I +GYTPIL CHTSH+ VKFA+L K D + +EK+P LKNGD ++KMIP +
Sbjct: 183 HPSQIEEGYTPILHCHTSHIPVKFAELIYKVDYCSHKIIEKRPPFLKNGDNCLIKMIPTE 362
Query: 336 PMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK 370
MVVE + P L RF V+D+ Q VAVGV ++VK+
Sbjct: 363 SMVVEPFSLSPPLSRFVVQDLHQIVAVGVAINVKR 467
>AL376457 similar to SP|P13549|EF10 Elongation factor 1-alpha somatic form
(EF-1-alpha-S). [African clawed frog] {Xenopus laevis},
partial (25%)
Length = 479
Score = 139 bits (350), Expect = 2e-33
Identities = 68/120 (56%), Positives = 84/120 (69%)
Frame = +2
Query: 270 RVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIV 329
+VI++NHP I GY P+LDCHT+H+A KFA+L K DR +G LE+ PK LK+G+A +V
Sbjct: 2 QVIVLNHPGQIGAGYAPVLDCHTAHIACKFAELLEKVDRRSGKTLEEAPKFLKSGEAAMV 181
Query: 330 KMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
KMIP KPMVVE +YP LGRFAVRDMRQTVAVGVI V+K + TK + K
Sbjct: 182 KMIPSKPMVVEKFADYPPLGRFAVRDMRQTVAVGVIKDVEKKAATSGKVTKSAATAAKGK 361
>TC91780 homologue to SP|P17786|EF1A_LYCES Elongation factor 1-alpha
(EF-1-alpha). [Tomato] {Lycopersicon esculentum},
partial (82%)
Length = 1165
Score = 129 bits (324), Expect = 2e-30
Identities = 60/89 (67%), Positives = 71/89 (79%)
Frame = +1
Query: 263 EAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLK 322
+ A FT++VII+NHP I GY P+LDCHTSH+AVKFA++ TK DR +G ELEK+PK LK
Sbjct: 763 DQASFTAQVIIMNHPGQIGNGYAPVLDCHTSHIAVKFAEILTKIDRRSGKELEKEPKFLK 942
Query: 323 NGDAGIVKMIPMKPMVVEGINEYPSLGRF 351
NGDAG+VKMIP KPMVVE EYP LG F
Sbjct: 943 NGDAGMVKMIPTKPMVVETFAEYPPLGSF 1029
Score = 105 bits (261), Expect = 4e-23
Identities = 59/96 (61%), Positives = 66/96 (68%), Gaps = 4/96 (4%)
Frame = +1
Query: 8 INIVFIGHVNSGKS----TTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKL 63
++IV IGH K T L +L + E KEAAEM RSFKYAWVLDKL
Sbjct: 4 LSIVVIGHCRLWKVDYPLVT*STSLVELTSVLLRGFE---KEAAEMNKRSFKYAWVLDKL 174
Query: 64 KAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
KAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 175KAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 282
Score = 82.4 bits (202), Expect = 3e-16
Identities = 39/47 (82%), Positives = 42/47 (88%)
Frame = +2
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKP 199
L A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGV+KP
Sbjct: 632 LLDALDQINEPKRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGVIKP 772
Score = 62.8 bits (151), Expect = 3e-10
Identities = 48/135 (35%), Positives = 63/135 (46%), Gaps = 4/135 (2%)
Frame = +3
Query: 25 GHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERERGIT---IDISMCKFE 81
GHL+YKLGGI K IE + + + + + L A + G T C E
Sbjct: 57 GHLIYKLGGIDKRVIERVREGSC*DEQEVIQVC-----LGA*QT*GRT*AWYHH*YCFVE 221
Query: 82 TTKYCCTL-IDAPGHRDFIN*EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCST 140
+ L D ++ EHDH L *LCC + H+ F SW +* WSD TC
Sbjct: 222 V*DH*VLLHCD*CPRTQGLHQEHDHWYLPG*LCCPHY*LHHWWF*SWYL*RWSDT*TCIA 401
Query: 141 CVHSWCEANDLLLQQ 155
C + WC+ NDLLL Q
Sbjct: 402 CFYPWCQTNDLLL*Q 446
Score = 34.7 bits (78), Expect = 0.074
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 3/77 (3%)
Frame = +2
Query: 290 CHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGI---VKMIPMKPMVVEGINEYP 346
C T+ + + L R TGV++ +SL + + ++ P ++ +
Sbjct: 836 CSTATLPTLLSSLLRS*PRLTGVQVRNLRRSLSS*RMVMQVWLR*FPPSLWLLRPLLSTH 1015
Query: 347 SLGRFAVRDMRQTVAVG 363
GRFAVRDMRQTVAVG
Sbjct: 1016HWGRFAVRDMRQTVAVG 1066
>TC85199 homologue to SP|O24534|EF1A_VICFA Elongation factor 1-alpha
(EF-1-alpha). [Broad bean] {Vicia faba}, partial (28%)
Length = 887
Score = 125 bits (314), Expect = 3e-29
Identities = 58/83 (69%), Positives = 68/83 (81%)
Frame = +3
Query: 263 EAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLK 322
EAA FTS+VII+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LK
Sbjct: 639 EAANFTSQVIIMNHPGQIGNGYAPVLDCHTSHIAVKFAELVTKIDRRSG*EIEKEPKFLK 818
Query: 323 NGDAGIVKMIPMKPMVVEGINEY 345
NGDAGI+KM+P KPMVVE EY
Sbjct: 819 NGDAGIIKMVPTKPMVVETFAEY 887
Score = 53.5 bits (127), Expect = 2e-07
Identities = 25/37 (67%), Positives = 29/37 (77%)
Frame = +2
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKS 37
M K +INIV IGHV+SGKSTT GHL+YKLGGI +
Sbjct: 506 MGKEKIHINIVVIGHVDSGKSTTTGHLIYKLGGIDNA 616
>TC84690 similar to GP|21540004|gb|AAM54368.1 elongation factor 1-alpha
{Trichophyton rubrum}, partial (24%)
Length = 546
Score = 125 bits (313), Expect = 4e-29
Identities = 61/109 (55%), Positives = 76/109 (68%), Gaps = 4/109 (3%)
Frame = +3
Query: 276 HPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMK 335
HP + GY P+L CHT+H+A KFA++ K DR TG E PK +K+GD+ IVKM+P K
Sbjct: 3 HPGQVGAGYAPVLVCHTAHIACKFAEIQEKIDRRTGKATEAAPKFIKSGDSAIVKMVPSK 182
Query: 336 PMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK----NGYKNKAATK 380
PM VE +YP LGRFAVRDMRQTVAVGVI +V+K +G K+A K
Sbjct: 183 PMCVEAFTDYPPLGRFAVRDMRQTVAVGVIKAVEKSVGGSGKVTKSAAK 329
>BG649063 homologue to PIR|F86214|F86 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 605
Score = 125 bits (313), Expect = 4e-29
Identities = 63/94 (67%), Positives = 74/94 (78%)
Frame = +2
Query: 142 VHSWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGM 201
V W + LL +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG +KPGM
Sbjct: 326 VLDWYKGPTLL--EALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGTMKPGM 499
Query: 202 VLTFAPTKLQSGVKSIQKFQENINEALPADIVGF 235
V+TFAPT LQ+ VKS++ E++ EALP D VGF
Sbjct: 500 VVTFAPTGLQTEVKSVEMHHESLTEALPRDHVGF 601
Score = 89.7 bits (221), Expect = 2e-18
Identities = 45/64 (70%), Positives = 48/64 (74%)
Frame = +2
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 152 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 331
Query: 61 DKLK 64
D K
Sbjct: 332 DWYK 343
>BF649588 similar to SP|P29521|EF11_ Elongation factor 1-alpha (EF-1-alpha).
[Carrot] {Daucus carota}, partial (18%)
Length = 599
Score = 120 bits (302), Expect = 8e-28
Identities = 60/84 (71%), Positives = 68/84 (80%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K INIV IGHV+SGKSTT GHL++KLG I KS IE+L KEAA+M RSFKYAWVL
Sbjct: 319 MAKKKVRINIVVIGHVDSGKSTTTGHLIHKLGCIHKSVIESLEKEAAKMNKRSFKYAWVL 498
Query: 61 DKLKAERERGITIDISMCKFETTK 84
DKLKAERERG+TIDI++ K ET K
Sbjct: 499 DKLKAERERGVTIDITLSKXETNK 570
>AJ499655 homologue to SP|P28295|EF1A_ Elongation factor 1-alpha
(EF-1-alpha). [Pin mould] {Absidia glauca}, partial
(18%)
Length = 275
Score = 104 bits (259), Expect = 8e-23
Identities = 55/89 (61%), Positives = 66/89 (73%), Gaps = 3/89 (3%)
Frame = +3
Query: 293 SHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFA 352
+ +A KFA+L K DR +G +LE PK +K+GDA IVKMIP KPM VE ++YP LGRFA
Sbjct: 6 TRIACKFAELQEKIDRRSGKKLEDNPKFVKSGDAAIVKMIPSKPMCVEAFSDYPPLGRFA 185
Query: 353 VRDMRQTVAVGVIMSVK---KNGYKNKAA 378
VRDMRQTVAVGVI SV+ K+G KAA
Sbjct: 186 VRDMRQTVAVGVIKSVEKVDKSGKTTKAA 272
>TC85202 homologue to GP|18873727|gb|AAL79775.1 elongation factor 1 alpha
{Saccharum hybrid cultivar CP72-2086}, partial (45%)
Length = 769
Score = 100 bits (250), Expect = 8e-22
Identities = 48/56 (85%), Positives = 51/56 (90%)
Frame = +2
Query: 44 KEAAEMKMRSFKYAWVLDKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
KEAAEM R+FKYAWVLDKLKAERERGITIDI++ KFETTKY CT IDAPGHRDFI
Sbjct: 263 KEAAEMNKRTFKYAWVLDKLKAERERGITIDIALWKFETTKYYCTDIDAPGHRDFI 430
Score = 87.4 bits (215), Expect = 1e-17
Identities = 58/154 (37%), Positives = 83/154 (53%), Gaps = 5/154 (3%)
Frame = +1
Query: 7 NINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKL--- 63
+INIV IGHV+SGKSTT GHL+YKLGGI K IE L + + + + +
Sbjct: 151 HINIVDIGHVDSGKSTTTGHLIYKLGGIDKRVIETLDEGSC*NEQTYIQVCMGS*QT*G* 330
Query: 64 --KAERERGITIDISMCKFETTKYCCTLIDAPGHRDFIN*EHDHRDLRS*LCCSSHQFHY 121
K R ++I + ++ C+ + EHD+ +L *LC + H H+
Sbjct: 331 A*KRNYHRYCLVEI*DHQVLLHRH*CS------RTS*FHQEHDYWNLPG*LCRAYH*LHH 492
Query: 122 SRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
F SW +*GW+D S+C++SW EA+DLLL Q
Sbjct: 493 WWF*SWYL*GWTDP*ARSSCIYSWSEADDLLL*Q 594
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,220,779
Number of Sequences: 36976
Number of extensions: 172245
Number of successful extensions: 894
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 851
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 869
length of query: 436
length of database: 9,014,727
effective HSP length: 99
effective length of query: 337
effective length of database: 5,354,103
effective search space: 1804332711
effective search space used: 1804332711
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135415.3


