

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135320.16 + phase: 0
(153 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
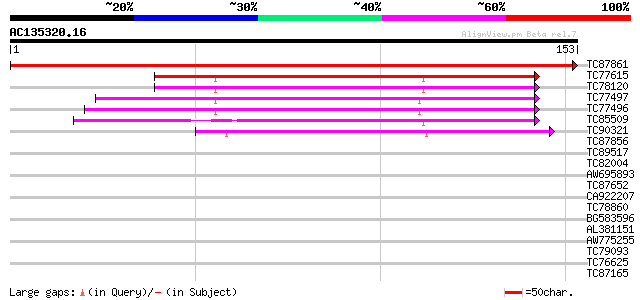
Score E
Sequences producing significant alignments: (bits) Value
TC87861 similar to PIR|B71412|B71412 ferredoxin [2Fe-2S] - Arabi... 312 4e-86
TC77615 similar to SP|P27788|FER3_MAIZE Ferredoxin III chloropl... 87 2e-18
TC78120 similar to PIR|G84673|G84673 probable ferredoxin [import... 87 3e-18
TC77497 similar to PIR|S62722|S62722 ferredoxin [2Fe-2S] fd1 pre... 82 8e-17
TC77496 similar to PIR|S62722|S62722 ferredoxin [2Fe-2S] fd1 pre... 82 1e-16
TC85509 similar to SP|P09911|FER1_PEA Ferredoxin I chloroplast ... 77 2e-15
TC90321 similar to GP|13174248|gb|AAK14422.1 putative ferredoxin... 61 2e-10
TC87856 similar to GP|19699067|gb|AAL90901.1 AT5g20250/F5O24_140... 28 1.9
TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein... 28 1.9
TC82004 similar to GP|9294687|dbj|BAB03053.1 gene_id:MHC9.8~unkn... 28 1.9
AW695893 homologue to GP|8489871|gb| DEAD box protein P68 {Pisum... 27 2.5
TC87652 similar to GP|13926327|gb|AAK49628.1 At1g26660/T24P13_4 ... 27 3.2
CA922207 similar to GP|10177923|dbj gb|AAF26028.1~gene_id:MCA23.... 27 3.2
TC78860 similar to GP|14030719|gb|AAK53034.1 AT4g32520/F8B4_220 ... 27 3.2
BG583596 similar to GP|4325376|gb| T3H13.12 gene product {Arabid... 27 4.2
AL381151 26 5.5
AW775255 26 5.5
TC79093 similar to GP|4102190|gb|AAD01431.1| 35 kDa seed maturat... 26 5.5
TC76625 similar to SP|O81363|RS4_PRUAR 40S ribosomal protein S4.... 26 5.5
TC87165 weakly similar to GP|21689637|gb|AAM67440.1 putative sto... 26 7.2
>TC87861 similar to PIR|B71412|B71412 ferredoxin [2Fe-2S] - Arabidopsis
thaliana, partial (66%)
Length = 695
Score = 312 bits (799), Expect = 4e-86
Identities = 153/153 (100%), Positives = 153/153 (100%)
Frame = +2
Query: 1 MATLQFTPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEG 60
MATLQFTPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEG
Sbjct: 44 MATLQFTPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEG 223
Query: 61 KTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDDVVERG 120
KTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDDVVERG
Sbjct: 224 KTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDDVVERG 403
Query: 121 YALLCASYPRSDCHIRVIPEDELLSMQLATAND 153
YALLCASYPRSDCHIRVIPEDELLSMQLATAND
Sbjct: 404 YALLCASYPRSDCHIRVIPEDELLSMQLATAND 502
>TC77615 similar to SP|P27788|FER3_MAIZE Ferredoxin III chloroplast
precursor (Fd III). [Maize] {Zea mays}, partial (65%)
Length = 906
Score = 87.4 bits (215), Expect = 2e-18
Identities = 44/106 (41%), Positives = 65/106 (60%), Gaps = 2/106 (1%)
Frame = +3
Query: 40 SSPSLTFTVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPA 98
SS S T ++YKV +I +GK + + D IL A D+G+++P+ C+ G C TC
Sbjct: 243 SSSSYRTTAMAAYKVKLIGPDGKENEFDAPDDVYILDAAEDAGVELPYSCRAGACSTCAG 422
Query: 99 RLISGTVDQSDG-MLSDDVVERGYALLCASYPRSDCHIRVIPEDEL 143
+++SG+VDQSDG L D+ ++ G+ L C SYP +DC I E EL
Sbjct: 423 KIVSGSVDQSDGSFLDDNQLKDGFVLTCVSYPTADCIIETHKEGEL 560
>TC78120 similar to PIR|G84673|G84673 probable ferredoxin [imported] -
Arabidopsis thaliana, partial (75%)
Length = 853
Score = 86.7 bits (213), Expect = 3e-18
Identities = 44/106 (41%), Positives = 64/106 (59%), Gaps = 2/106 (1%)
Frame = +1
Query: 40 SSPSLTFTVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPA 98
SS S T ++YKV +I +GK + E D IL A ++G+++P+ C+ G C TC
Sbjct: 235 SSASSRVTAVAAYKVKLIGPDGKENEFEATDDTYILDAAENAGVELPYSCRAGACSTCAG 414
Query: 99 RLISGTVDQSDG-MLSDDVVERGYALLCASYPRSDCHIRVIPEDEL 143
+++SG+VDQSDG L D+ + GY L C +YP SDC I E +L
Sbjct: 415 KVVSGSVDQSDGSFLDDNQLNEGYVLTCVAYPTSDCVIHTHKEGDL 552
>TC77497 similar to PIR|S62722|S62722 ferredoxin [2Fe-2S] fd1 precursor
non-photosynthetic - sweet orange, partial (84%)
Length = 880
Score = 82.0 bits (201), Expect = 8e-17
Identities = 43/122 (35%), Positives = 67/122 (54%), Gaps = 2/122 (1%)
Frame = +2
Query: 24 PSQLNTRPRLGSGSHPSSPSLTFTVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGL 82
PS L + + S S + + YKV +I +G + + D IL A D+G+
Sbjct: 212 PSSLRSVKNVSKTFGLKSSSFRVSAMAVYKVKLIGPDGTENEFDAPDDSYILDSAEDAGV 391
Query: 83 DVPHDCKLGVCMTCPARLISGTVDQSD-GMLSDDVVERGYALLCASYPRSDCHIRVIPED 141
++P+ C+ G C TC ++++G+VDQSD L + +E+GY L C SYP+SD I E+
Sbjct: 392 ELPYSCRAGACSTCAGQVVTGSVDQSDQSFLDEQQIEKGYLLTCVSYPKSDTVIYTHKEE 571
Query: 142 EL 143
EL
Sbjct: 572 EL 577
>TC77496 similar to PIR|S62722|S62722 ferredoxin [2Fe-2S] fd1 precursor
non-photosynthetic - sweet orange, partial (84%)
Length = 827
Score = 81.6 bits (200), Expect = 1e-16
Identities = 44/125 (35%), Positives = 68/125 (54%), Gaps = 2/125 (1%)
Frame = +2
Query: 21 LPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKV-VIEHEGKTTQLEVEPDETILSKALD 79
L PS L + + S S + + YKV +I+ +G + + D IL A +
Sbjct: 146 LKNPSTLRSVKNVSKRFGLKSSSFRISAMAVYKVKLIQPDGTENEFDAPDDYYILDSAEE 325
Query: 80 SGLDVPHDCKLGVCMTCPARLISGTVDQSD-GMLSDDVVERGYALLCASYPRSDCHIRVI 138
+G+++P+ C+ G C TC +++SG+VDQSD L +E+GY L C SYP+SD I
Sbjct: 326 AGVELPYSCRAGACSTCAGQVVSGSVDQSDQSFLDKQQIEKGYLLTCVSYPKSDTVIYTH 505
Query: 139 PEDEL 143
E+EL
Sbjct: 506 KEEEL 520
>TC85509 similar to SP|P09911|FER1_PEA Ferredoxin I chloroplast precursor.
[Garden pea] {Pisum sativum}, partial (90%)
Length = 831
Score = 77.4 bits (189), Expect = 2e-15
Identities = 43/127 (33%), Positives = 66/127 (51%), Gaps = 1/127 (0%)
Frame = +2
Query: 18 TTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQLEVEPDETILSKA 77
TT +PS + + G + T+ V+ ++ EG T + + D IL A
Sbjct: 200 TTTKAFPSGFGLKSKTGKRGDLAVAMATYKVK-----LVTPEG-TQEFDCPSDVYILDHA 361
Query: 78 LDSGLDVPHDCKLGVCMTCPARLISGTVDQSDG-MLSDDVVERGYALLCASYPRSDCHIR 136
+ G+D+P+ C+ G C +C ++ +G VDQSDG L DD +E G+ L C +YP SD I
Sbjct: 362 EEVGIDLPYSCRAGSCSSCAGKVAAGAVDQSDGSFLDDDQIEEGWVLTCVAYPTSDVTIE 541
Query: 137 VIPEDEL 143
E+EL
Sbjct: 542 THKEEEL 562
>TC90321 similar to GP|13174248|gb|AAK14422.1 putative ferredoxin {Oryza
sativa}, partial (65%)
Length = 649
Score = 60.8 bits (146), Expect = 2e-10
Identities = 32/100 (32%), Positives = 54/100 (54%), Gaps = 3/100 (3%)
Frame = +3
Query: 51 SYKVVIE--HEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQS 108
++KV + G + V D+ IL A + +P C+ G C +C R+ +G + Q
Sbjct: 216 THKVTVHDRQRGVVHEFLVPEDQYILHTAESQNITLPFACRHGCCTSCAVRIKNGKIKQP 395
Query: 109 DGM-LSDDVVERGYALLCASYPRSDCHIRVIPEDELLSMQ 147
+ + +S ++ E+GYALLC S+P SD + EDE+ +Q
Sbjct: 396 EALGISAELREQGYALLCVSFPYSDLEVETQDEDEVYWLQ 515
>TC87856 similar to GP|19699067|gb|AAL90901.1 AT5g20250/F5O24_140
{Arabidopsis thaliana}, partial (45%)
Length = 1402
Score = 27.7 bits (60), Expect = 1.9
Identities = 12/26 (46%), Positives = 14/26 (53%)
Frame = -2
Query: 9 LTLFRQKHPTTKLPYPSQLNTRPRLG 34
LT+ KHPTT + P TRP G
Sbjct: 684 LTIIHSKHPTTSIHVPYLKQTRPITG 607
>TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein AT4g40080
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1177
Score = 27.7 bits (60), Expect = 1.9
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Frame = -2
Query: 17 PTTKLPYPSQLNTRPRLGSGSHPSSPSL---TFTVRSSYKVV 55
P++ P+PS L+ PR+ S P S TF + S +K +
Sbjct: 921 PSSSQPFPSILSPNPRISSPQPPPSSLFSLCTFPISSKHKPI 796
>TC82004 similar to GP|9294687|dbj|BAB03053.1 gene_id:MHC9.8~unknown
protein {Arabidopsis thaliana}, partial (50%)
Length = 603
Score = 27.7 bits (60), Expect = 1.9
Identities = 13/40 (32%), Positives = 18/40 (44%)
Frame = +2
Query: 22 PYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGK 61
P P NT+P G P+ T V +++ HEGK
Sbjct: 59 PMPKLRNTKPNSGHYEERPVPAGTLNVAQLRHIILLHEGK 178
>AW695893 homologue to GP|8489871|gb| DEAD box protein P68 {Pisum sativum},
partial (21%)
Length = 646
Score = 27.3 bits (59), Expect = 2.5
Identities = 16/44 (36%), Positives = 21/44 (47%)
Frame = +3
Query: 1 MATLQFTPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSL 44
++ L PL LF P KLP+ + S SH +SPSL
Sbjct: 141 LSHLHNLPLHLFLLPWPMAKLPHHHHHHNNNLAFSNSHSNSPSL 272
>TC87652 similar to GP|13926327|gb|AAK49628.1 At1g26660/T24P13_4
{Arabidopsis thaliana}, partial (83%)
Length = 1081
Score = 26.9 bits (58), Expect = 3.2
Identities = 17/54 (31%), Positives = 27/54 (49%), Gaps = 2/54 (3%)
Frame = +1
Query: 14 QKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLT--FTVRSSYKVVIEHEGKTTQL 65
+K TT+L +PSQ S H S PSL S ++ I++ +T++L
Sbjct: 73 EKRTTTQLHHPSQQQQHSCSCSDLHQSQPSLARISLPSSDVELAIDNSDRTSEL 234
>CA922207 similar to GP|10177923|dbj gb|AAF26028.1~gene_id:MCA23.21~similar
to unknown protein {Arabidopsis thaliana}, partial (21%)
Length = 792
Score = 26.9 bits (58), Expect = 3.2
Identities = 18/65 (27%), Positives = 30/65 (45%)
Frame = -1
Query: 82 LDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDDVVERGYALLCASYPRSDCHIRVIPED 141
L+V +DC +C R +GT+ +DG +SD V C + R + + +D
Sbjct: 417 LNVQNDCNF-LC**QAYRESTGTISTTDGSISDPVSAAEENAFCKACARFGLGLYLYHDD 241
Query: 142 ELLSM 146
+ SM
Sbjct: 240 QTASM 226
>TC78860 similar to GP|14030719|gb|AAK53034.1 AT4g32520/F8B4_220
{Arabidopsis thaliana}, partial (33%)
Length = 915
Score = 26.9 bits (58), Expect = 3.2
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 10/53 (18%)
Frame = +3
Query: 81 GLDVPHDCKLGVCMTCPARLISGT----------VDQSDGMLSDDVVERGYAL 123
GLD+PH L R +SGT +D+S G++ D++E+ AL
Sbjct: 732 GLDLPHGGHLSHGFMTAKRRVSGTSIYFESMPYRLDESTGVIDYDMLEKTAAL 890
>BG583596 similar to GP|4325376|gb| T3H13.12 gene product {Arabidopsis
thaliana}, partial (46%)
Length = 778
Score = 26.6 bits (57), Expect = 4.2
Identities = 15/43 (34%), Positives = 18/43 (40%)
Frame = -2
Query: 7 TPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVR 49
T LT HP +P PS + + S HP SP F R
Sbjct: 777 THLTNSSSSHPQFPIPNPSHASQEK*ISS*HHPHSP*TPFQSR 649
>AL381151
Length = 491
Score = 26.2 bits (56), Expect = 5.5
Identities = 13/24 (54%), Positives = 14/24 (58%)
Frame = -3
Query: 121 YALLCASYPRSDCHIRVIPEDELL 144
YA AS R DCH R P+D LL
Sbjct: 264 YAKEIASCSRHDCHHRHCPQDVLL 193
>AW775255
Length = 310
Score = 26.2 bits (56), Expect = 5.5
Identities = 12/20 (60%), Positives = 12/20 (60%)
Frame = -2
Query: 6 FTPLTLFRQKHPTTKLPYPS 25
F PL RQKH KL YPS
Sbjct: 216 FRPLDYHRQKHNEWKLHYPS 157
>TC79093 similar to GP|4102190|gb|AAD01431.1| 35 kDa seed maturation protein
{Glycine max}, partial (57%)
Length = 1127
Score = 26.2 bits (56), Expect = 5.5
Identities = 12/28 (42%), Positives = 14/28 (49%)
Frame = -3
Query: 16 HPTTKLPYPSQLNTRPRLGSGSHPSSPS 43
HP PY + P+L S HPS PS
Sbjct: 726 HPLPASPYQQHSSYTPQLCSPLHPSLPS 643
>TC76625 similar to SP|O81363|RS4_PRUAR 40S ribosomal protein S4. [Apricot]
{Prunus armeniaca}, partial (13%)
Length = 1172
Score = 26.2 bits (56), Expect = 5.5
Identities = 14/40 (35%), Positives = 16/40 (40%)
Frame = -3
Query: 3 TLQFTPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSP 42
T+ PL L R H LPY T +G HP P
Sbjct: 912 TVNLIPLPLGRDTHGFVPLPYNIPHQTVQEQKNGEHPHLP 793
>TC87165 weakly similar to GP|21689637|gb|AAM67440.1 putative storage
protein {Arabidopsis thaliana}, partial (71%)
Length = 1237
Score = 25.8 bits (55), Expect = 7.2
Identities = 13/29 (44%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Frame = +2
Query: 37 SHPSSPSLTFTVRSSYKVVIEHEG-KTTQ 64
+HP S S TF +++ +V I+HE K TQ
Sbjct: 53 NHPLSSSSTFIIQTFLQVQIKHEAPKNTQ 139
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,562,924
Number of Sequences: 36976
Number of extensions: 55824
Number of successful extensions: 471
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 413
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 467
length of query: 153
length of database: 9,014,727
effective HSP length: 88
effective length of query: 65
effective length of database: 5,760,839
effective search space: 374454535
effective search space used: 374454535
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC135320.16


