

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
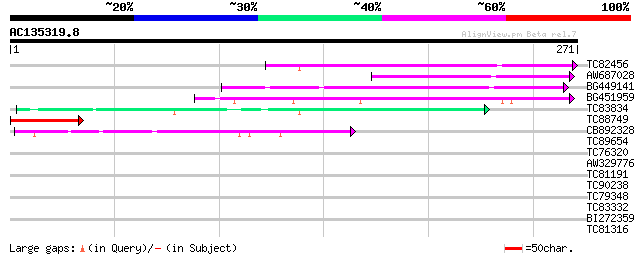
Query= AC135319.8 - phase: 0 /pseudo
(271 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 61 5e-10
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 60 7e-10
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 51 4e-07
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 49 3e-06
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 47 6e-06
TC88749 44 5e-05
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 44 7e-05
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 40 0.001
TC76320 homologue to SP|P56337|IF55_SOLTU Initiation factor 5A-5... 29 2.2
AW329776 28 3.7
TC81191 similar to GP|6143879|gb|AAF04426.1| unknown protein {Ar... 27 6.4
TC90238 similar to PIR|T07126|T07126 magnesium chelatase (EC 4.9... 27 6.4
TC79348 weakly similar to GP|1546698|emb|CAA67340.1 peroxidase A... 27 8.3
TC83332 weakly similar to GP|13491750|gb|AAK27968.1 cysteine pro... 27 8.3
BI272359 similar to GP|16604495|gb AT4g20170/F1C12_90 {Arabidops... 27 8.3
TC81316 similar to PIR|C96704|C96704 unknown protein 23065-2035... 27 8.3
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 60.8 bits (146), Expect = 5e-10
Identities = 34/150 (22%), Positives = 68/150 (44%), Gaps = 1/150 (0%)
Frame = +2
Query: 123 RGDLTEMQYLISKLE-ENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTY 181
RGD+ + +++ +N + + + + +Q++FW + F V+ +D+TY
Sbjct: 710 RGDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTY 889
Query: 182 KTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVT 241
T+ Y +PL VGV T + + ++ TW L+ + ++ P ++T
Sbjct: 890 LTSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHA--PNGIIT 1063
Query: 242 NRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
D +M A+ A P + C +H+ K V
Sbjct: 1064EEDKAMKNAIEVAFPKARHRWCLWHIMKKV 1153
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 60.5 bits (145), Expect = 7e-10
Identities = 34/97 (35%), Positives = 49/97 (50%)
Frame = +1
Query: 174 VLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNS 233
VL D+TYK N Y PL G T + + E +++ W L L+ +E +
Sbjct: 4 VLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECME--N 177
Query: 234 DMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGKN 270
PK VVT+ D SM +A+ PD+S LC +H+ KN
Sbjct: 178 KFPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKN 288
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 51.2 bits (121), Expect = 4e-07
Identities = 32/166 (19%), Positives = 82/166 (49%)
Frame = +3
Query: 102 KESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEENGYVHYVREKPESQTVQDIFWTH 161
++ + N+ Q + + ++T DL M I + N Y + + +++I W++
Sbjct: 177 EKDVRNLLQSFRKLDPEEETL--DLLRMCRNIKDKDPNFKFEYTLDA--NNRLENIAWSY 344
Query: 162 PTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWA 221
+S++L++ F ++ D+T++ + MPL VG+ + + + E +F+WA
Sbjct: 345 ASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSFSWA 524
Query: 222 LQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
++ L + N P+ ++T+++ + +A++ +P + C + +
Sbjct: 525 IKAFLGFM--NGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMI 656
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 48.5 bits (114), Expect = 3e-06
Identities = 48/192 (25%), Positives = 77/192 (40%), Gaps = 10/192 (5%)
Frame = +2
Query: 89 KPKNILTNLKKKRKESIT-NIKQVYNERHKFKKTKRGDLTEMQYLIS------KLEENGY 141
KPK IL + R IT + KQ + + RG E L+ K G
Sbjct: 77 KPKEILEEIH--RVHGITLSYKQAWRGKEHIMAAMRGSFEEGYRLLPQYCAHVKRTNPGS 250
Query: 142 VHYVREKPESQTVQDIFWTHPTSVK-LFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTY 200
+ V P Q +F + S+ L N +L +D Y + Y L G
Sbjct: 251 IASVYGNPSDNCFQRLFISFQASIYGLLNACRPLLGLDRIYLKSKYLGTLLLATGFDGDG 430
Query: 201 LTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSD-MPKV-VVTNRDPSMMKAVTNALPDS 258
+ + F + E +DN+ W L L LLE N++ MP++ ++++R ++ V P +
Sbjct: 431 ALFPLAFGVVDEENDDNWMWFLSKLHNLLEINTENMPRLTILSDRQQGIVDGVEANFPTA 610
Query: 259 SAILCYFHVGKN 270
C H+ N
Sbjct: 611 FHGFCMRHLSDN 646
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 47.4 bits (111), Expect = 6e-06
Identities = 45/232 (19%), Positives = 90/232 (38%), Gaps = 6/232 (2%)
Frame = +3
Query: 4 LVCERSGEHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYV 63
L C G +TK + + + GC +R +V E+ W++ + HNH + +
Sbjct: 351 LCCSSQG---FKRTKDVNNLRKETRTGCPAMIRMKLV-ESQRWRICEVTLEHNHVLGAKI 518
Query: 64 AGHLLAGRLMEDDK-----KIVHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNERHKF 118
+ L D K+ H L N+ +N + R S Y+ +
Sbjct: 519 HKSIKKNSLPSSDAEGKTIKVYHALVIDTEGNDNLNSNARDDRAFS------KYSNKLNL 680
Query: 119 KKTKRGDLTEMQYLISKLE-ENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIM 177
+K GD + + +++ N Y+ + + +++ W S F V+
Sbjct: 681 RK---GDTQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYF 851
Query: 178 DSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLL 229
D+TY N Y +PL +VG+ + + + E +++ W + +K +
Sbjct: 852 DNTYLVNKYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCI 1007
>TC88749
Length = 1221
Score = 44.3 bits (103), Expect = 5e-05
Identities = 20/35 (57%), Positives = 26/35 (74%)
Frame = +2
Query: 1 MLELVCERSGEHKLPKTKVKHEATESRKCGCLFKV 35
MLEL+CERSG+HK+ K ++KHEAT SR +V
Sbjct: 647 MLELMCERSGDHKVSKKRLKHEAT*SRNSAVYVQV 751
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 43.9 bits (102), Expect = 7e-05
Identities = 45/174 (25%), Positives = 75/174 (42%), Gaps = 11/174 (6%)
Frame = +1
Query: 3 ELVCERSG-EHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVR 61
+ C + G ++ P +V ++ +SR CL VR V +E WK+T L HNHE V
Sbjct: 292 DFYCSKQGFKNNEPDGEVAYKRADSRT-NCLAMVRFNVTKEG-VWKVTKLILDHNHEFVP 465
Query: 62 YVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNI-------KQVYN- 113
+LL R M + + D SLV +TN+ + + + K ++N
Sbjct: 466 LQQRYLL--RSMRNMSNLKEDRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMHNY 639
Query: 114 -ERHKFKKTKRGDLTE-MQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSV 165
K K + GD + +L SK ++ +Y + + ++FW SV
Sbjct: 640 VSTGKLKFIEAGDAQSLLNHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDGKSV 801
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 40.0 bits (92), Expect = 0.001
Identities = 21/67 (31%), Positives = 29/67 (42%)
Frame = +2
Query: 129 MQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 188
+ YL EN Y + +IFW T ++ F +I D+TYKTN YR+
Sbjct: 134 LDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTNQYRV 313
Query: 189 PLFEIVG 195
P G
Sbjct: 314 PFASFTG 334
>TC76320 homologue to SP|P56337|IF55_SOLTU Initiation factor 5A-5 (eIF-5A 5)
(eIF-4D). [Potato] {Solanum tuberosum}, complete
Length = 715
Score = 28.9 bits (63), Expect = 2.2
Identities = 19/74 (25%), Positives = 32/74 (42%), Gaps = 4/74 (5%)
Frame = +3
Query: 6 CERSGEHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKL----TILNGVHNHEMVR 61
C+ PK ++H+ K G + K G++V +N K+ T G H H
Sbjct: 42 CQTRNTILSPKQMLEHQRHIHNKPGTIRK-NGHIVIKNRPCKVVEVSTSKTGKHGHAKCH 218
Query: 62 YVAGHLLAGRLMED 75
+V + G+ +ED
Sbjct: 219 FVGIDIFTGKKLED 260
>AW329776
Length = 653
Score = 28.1 bits (61), Expect = 3.7
Identities = 19/71 (26%), Positives = 30/71 (41%)
Frame = +3
Query: 100 KRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEENGYVHYVREKPESQTVQDIFW 159
KR+ I N KQ KK + GD+ ++++L + Y H V + T W
Sbjct: 48 KRQHEIQNTKQS-------KKGRSGDINKVRHLDGSRRADEYQHQVAKSSTQAT-----W 191
Query: 160 THPTSVKLFNT 170
TS + F +
Sbjct: 192 NESTSSRAFRS 224
>TC81191 similar to GP|6143879|gb|AAF04426.1| unknown protein {Arabidopsis
thaliana}, partial (98%)
Length = 1067
Score = 27.3 bits (59), Expect = 6.4
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = +1
Query: 15 PKTKVKHEATESRKCGCLFKVRGYVVRENNAW 46
PKT + EAT+ R C+ + GY+ + N W
Sbjct: 1 PKTTMT-EATQPRTIICIGDIHGYITKLQNLW 93
>TC90238 similar to PIR|T07126|T07126 magnesium chelatase (EC 4.99.1.-)
chain chlH - soybean, partial (15%)
Length = 844
Score = 27.3 bits (59), Expect = 6.4
Identities = 12/46 (26%), Positives = 23/46 (49%)
Frame = -1
Query: 165 VKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFM 210
+ +N +++M+ Y NLY PLF + +T L +S + +
Sbjct: 160 LSFYNKEKVLVMMERWYLRNLYTFPLFFQICLTKVELCFSTQLNLV 23
>TC79348 weakly similar to GP|1546698|emb|CAA67340.1 peroxidase ATP3a
{Arabidopsis thaliana}, partial (37%)
Length = 1027
Score = 26.9 bits (58), Expect = 8.3
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = +3
Query: 159 WTHPTSVKLFNTFPTVLIMDSTYKT 183
W HP+S +F T P +MD Y T
Sbjct: 264 WHHPSSACIFMTVPLEDVMDQFYLT 338
>TC83332 weakly similar to GP|13491750|gb|AAK27968.1 cysteine protease
{Ipomoea batatas}, partial (23%)
Length = 422
Score = 26.9 bits (58), Expect = 8.3
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 7/53 (13%)
Frame = +3
Query: 98 KKKR----KESITNIK---QVYNERHKFKKTKRGDLTEMQYLISKLEENGYVH 143
K+KR KE++ I+ V N+ +K DLT +++ ++ + NGYVH
Sbjct: 237 KQKRFGIFKENVNYIEAFNNVGNKSYKLGLNHFADLTNHEFIAARNKFNGYVH 395
>BI272359 similar to GP|16604495|gb AT4g20170/F1C12_90 {Arabidopsis
thaliana}, partial (39%)
Length = 724
Score = 26.9 bits (58), Expect = 8.3
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = -1
Query: 66 HLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIK-QVYNERHK 117
HLL +L K+I H +L + NIL N+KK+ S I Q + HK
Sbjct: 526 HLLNCKLRVFRKRIKHSFNSTLWRHINILINIKKEHPFSHKLISMQTIIQNHK 368
>TC81316 similar to PIR|C96704|C96704 unknown protein 23065-20358
[imported] - Arabidopsis thaliana, partial (25%)
Length = 814
Score = 26.9 bits (58), Expect = 8.3
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = -3
Query: 141 YVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMP 189
++H +RE P + + + T+V L FP +LI ST +Y P
Sbjct: 422 FIHSIREMPSATAIPIVSEII*TTVLLSVVFPLLLISASTPAMAMYINP 276
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,191,119
Number of Sequences: 36976
Number of extensions: 102105
Number of successful extensions: 593
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 590
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 592
length of query: 271
length of database: 9,014,727
effective HSP length: 95
effective length of query: 176
effective length of database: 5,502,007
effective search space: 968353232
effective search space used: 968353232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135319.8


